Total Solution for MGI Platforms
Background
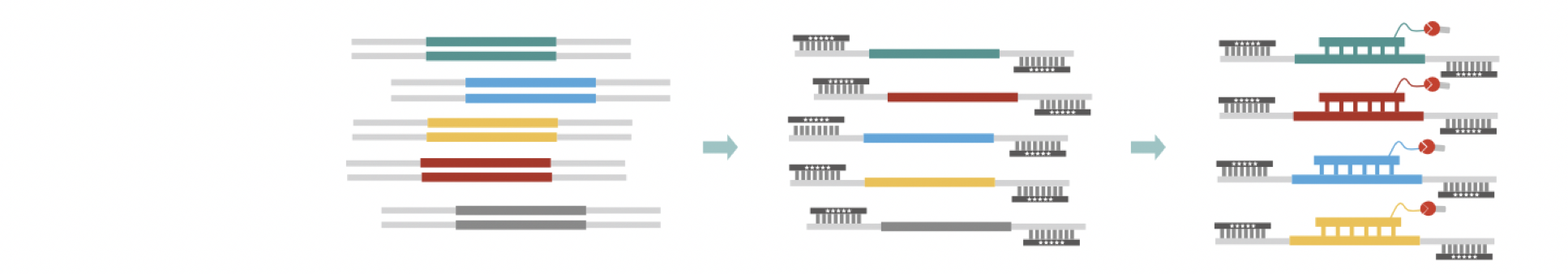
Nanodigmbio provides NGS total solution on MGI platforms, including library preparation and hybridization capture.Solution
|
|
Library preparation |
Blocking |
Target capture |
|
Exome sequencing |
|||
|
To learn more, visit Whole Exome Sequencing solution |
|||
|
Solid tumor analysis |
NanOnCT Panel |
||
|
Hematologic malignancy analysis |
|||
|
Low-frequency mutation analysis |
|||
|
To learn more, visit Low-frequency Mutation Analysis solution |
|||
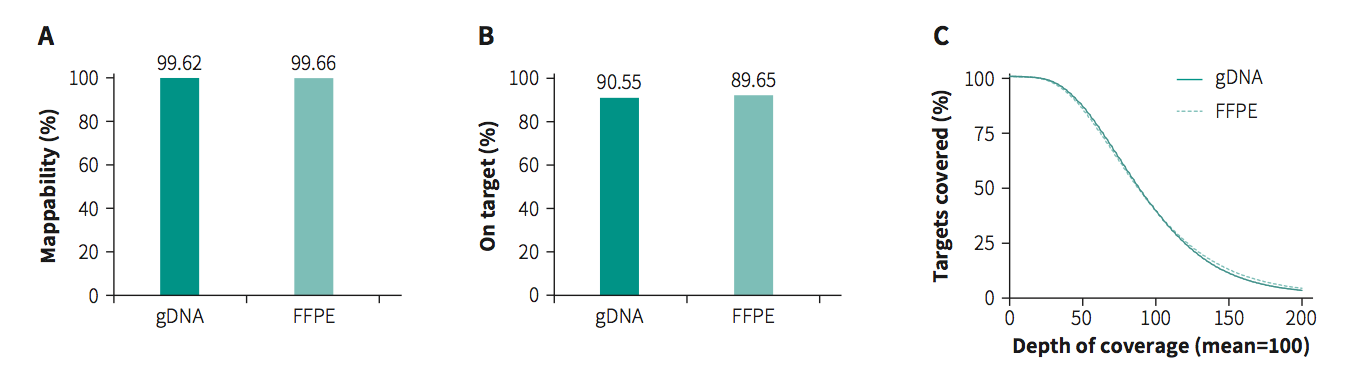
Fig 1. Capture performance of exome sequencing. The gDNA and FFPE libraries were prepared using NadPrep library kits for MGI. Exome Plus Panel v2.0 was used for target enrichment. The enriched libraries were sequenced on DNBSEQ-G400 (PE150) and subsampled to 35 M read pairs for the analysis of A. mappablity, B. on-target rate and C. coverage uniformity.
Table 1. Performance of the NanOnCT Panel v1.0
|
|
Mappability (%) |
On-target rate (%) |
Duplication (%) |
|
gDNA-1 |
99.75 |
87.32 |
2.91 |
|
gDNA-2 |
99.81 |
85.28 |
2.91 |
Solutions
- Methyl Library Preparation Total Solution
- Sequencing single library on different platform--Universal Stubby Adapter (UDI)
- HRD score Analysis
- Unique Dual Index for MGI platforms
- RNA-Cap Sequencing of Human Respiratory Viruses Including SARS-CoV-2
- Total Solution for RNA-Cap Sequencing
- Total Solution for MGI Platforms
- Whole Exome Sequencing
- Low-frequency Mutation Analysis
Events
-
Exhibition Preview | Nanodigmbio Invites You to Join Us at Hospitalar 2025, Brazil International Medical Device Exhibition in São Paulo

-
Exhibition Preview | Nanodigmbio invites you to join us at Denver 2024 Annual Meeting of the American Society of Human Genetics (ASHG)

-
Exhibition Preview | Nanodigmbio invites you to join us at Sapporo 2024 Annual Meeting of the Japan Society of Human Genetics (JSHG)

-
Exhibition Preview | Nanodigmbio invites you to join us at Association for Diagnostics & Laboratory Medicine (ADLM)

-
Exhibition Preview | Nanodigmbio invites you to join us at Medlab Asia & Asia Health 2024

-
Exhibition Preview | Nanodigmbio invites you to the Annual Meeting of the European Association for Cancer Research (EACR) 2024 in the Netherlands


