
NadPrep RNA & DNA Library Co-Preparation Module
#1002412
- Product details
- Kit Content
- FAQ
- Ordering Information
- Resources
Feature
● Flexible Compatibility
Supports mixed samples with varying ratios of 10-100 ng RNA and 1-100 ng DNA
Compatible with both MGI and Illumina sequencing platforms
● Simple and Rapid
Single-tube operation, quick and convenient
3-3.5 hrRNA & DNA library co-preparation in a single process in 3-3.5 hours
● Efficiency and Uniformity
High efficiency and stability of library yield
Uniform size distribution of fragment
Low GC bias
● Background Pathogenic Microorganism Control
Comprehensive control of background contamination of pathogenic microorganisms throughout the entire process, including environmental control, consumables control, raw material control, process control, manufacturing process control, quality
control, etc.
Workflow
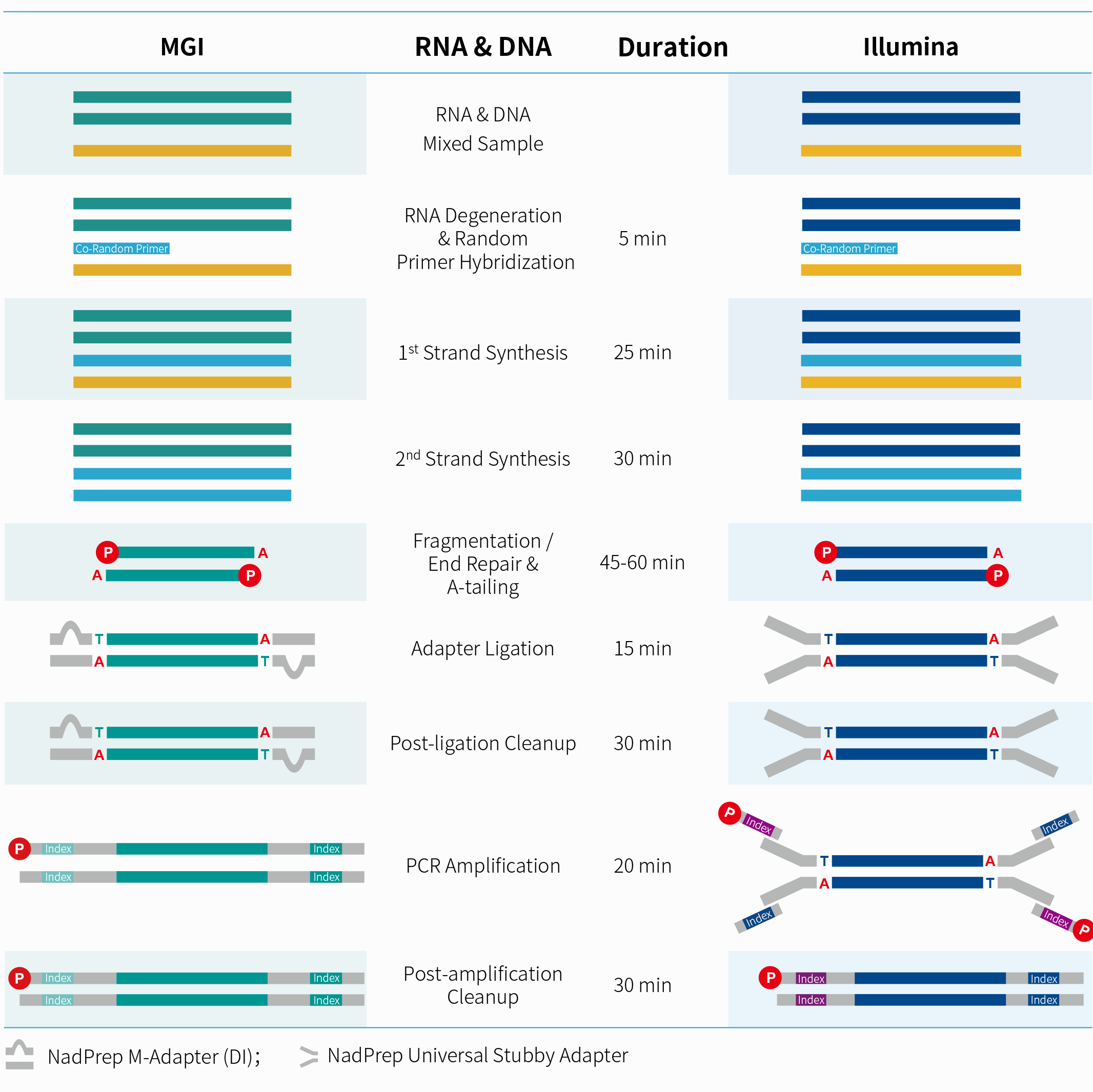
Performance
Flexible Compatibility
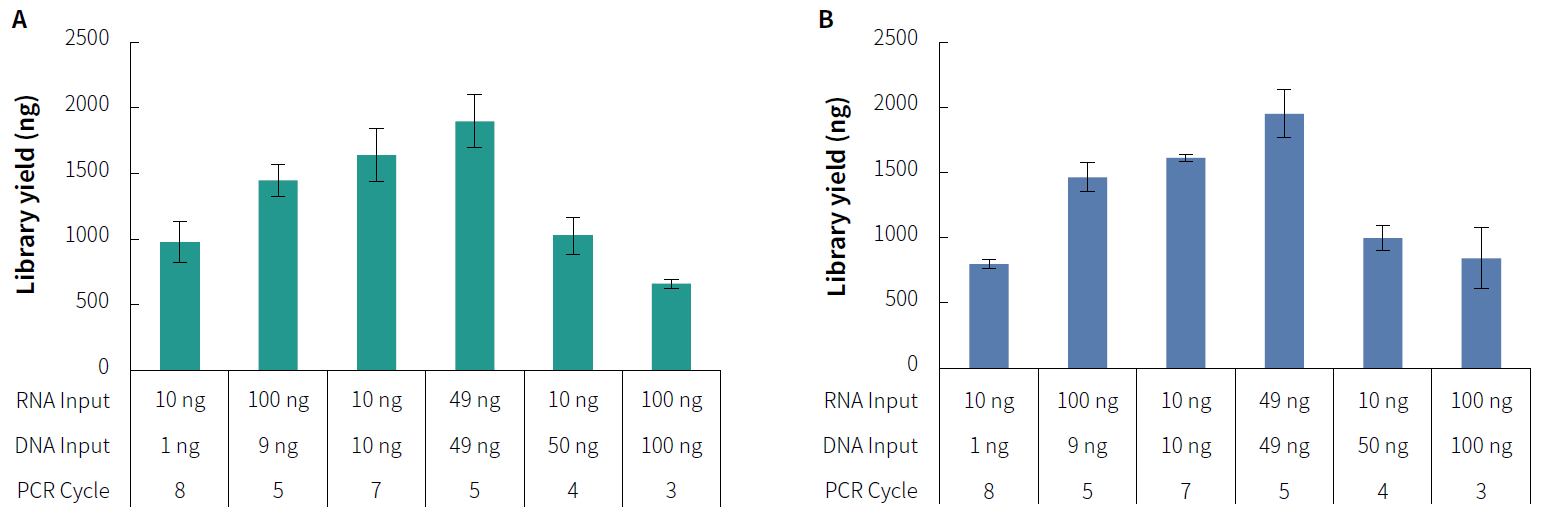
Figure 1. NadPrep RNA & DNA Library Co-Preparation Module for library co-preparation with mixed RNA & DNA samples in varying initial ratios. A. Library
yield (MDI); B. Library yield (UDI). Utilize NadPrep RNA & DNA Library Co-Preparation Module in conjunction with NadPrep Universal Adapter (MDI) Module (for MGI)
and NadPrep Universal Stubby Adapter (UDI) Module for library co-preparation.
Note: The RNA samples are from K562 cell lines; the DNA samples are Human Genomic DNA Standards (Promega, G1521).
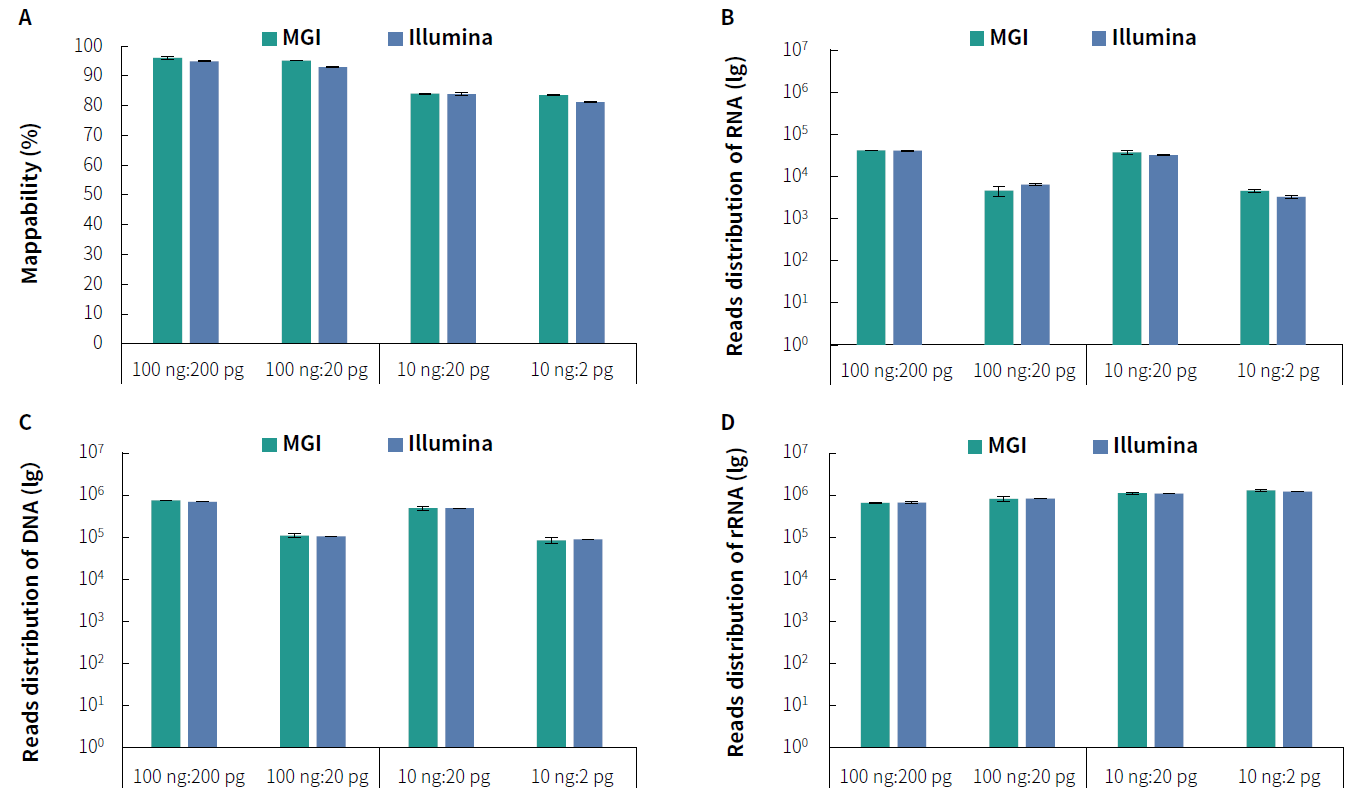
Figure 2. Detection of tNGS Total Solution with NadPrep RNA & DNA co-preparation on dual sequencing platforms. A. Mappability; B. Reads distribution
of Coronavirus; C. Reads distribution of Candida albicans; D. Reads distribution of rRNA. Pre-libraries preparation were performed using NadPrep RNA & DNA Library Co-Preparation Module. Hybridization capture were completed with NadPrep ES Hybrid Capture Reagents and NEX-t Panel v1.0, followed by sequencing on
DNBSEQ-T7, PE150 and Illumina NovaSeq 6000, PE150. For each sample, 0.5 Gb was used for data analysis.
Note: The scale on the X-axis represents the proportional initial input amounts of the host-mimicking mixed nucleic acid samples to the input amounts of the pathogenic mixed nucleic acid samples. The host-mimicking mixed nucleic acid samples consist of Human Brain Total RNA Standard (Clontech, 636530) and Human
Genomic DNA Standard (Promega, G1521) mixed at a 1:1 ratio. The pathogenic mixed nucleic acid samples consist of the Reference material for SARS-CoV-2 Omicron Variant Genomic RNA (National Institute of Metrology, NIM-RM 5225; 1 pg equals 50 copies) and Candida albicans standard (ATCC, 10231; 1 pg equals 6000 copies)
mixed at a 1:1 ratio.
Higher Sensitivity
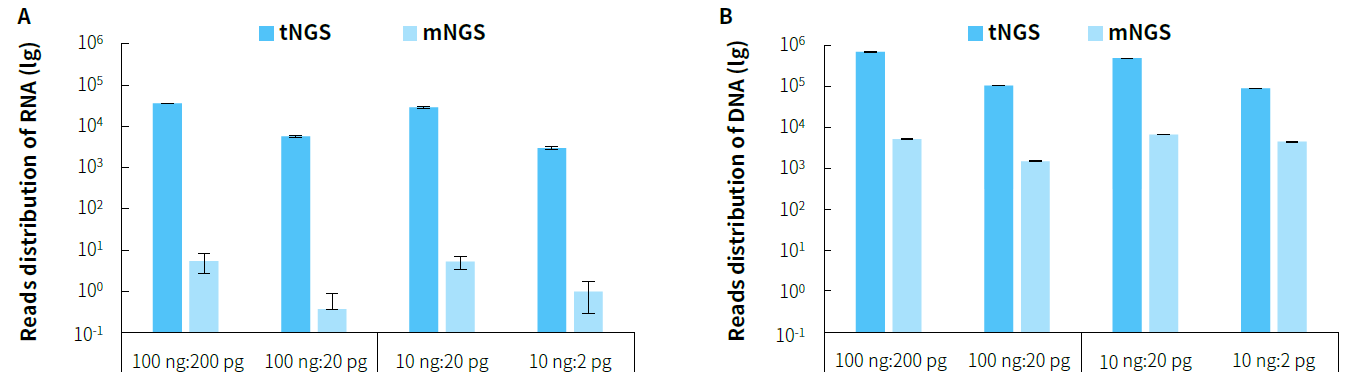
Figure 3. Distribution of detected reads in simulated pathogenic microbial samples using tNGS and mNGS based on the NadPrep RNA & DNA Library CoPreparation Module. A. Reads distribution of Coronavirus; B. Reads distribution of Candida albicans.
Note: Sequencing platform: Illumina NovaSeq 6000, PE150.
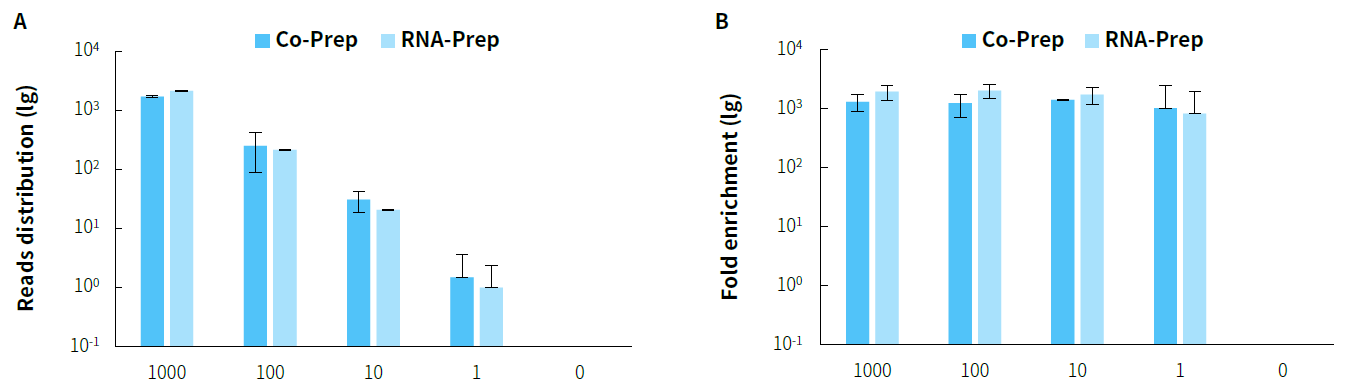
Figure 4. Capture performance of mixed RNA samples in varying copy numbers with co-preparation and RNA-preparation. A. Reads distribution; B. Enrichment fold.
Note: The X-axis represents the copy number of the Reference material for SARS-CoV-2 Omicron Variant Genomic RNA. The samples are derived from a mixture of
Human Brain Total RNA Standard (Clontech, 636530) and Omicron virus samples in varying copy numbers.
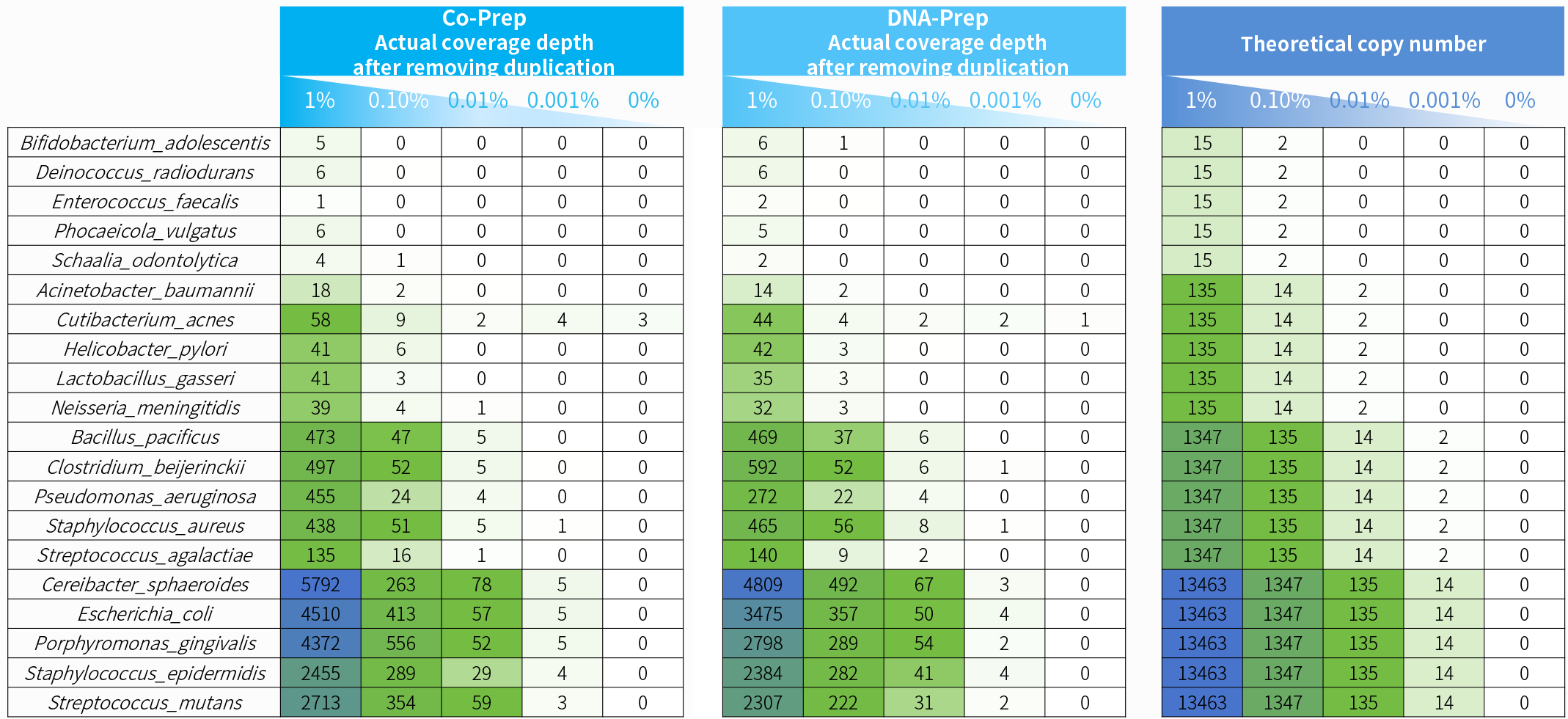
Figure 5. Captured coverage depth after duplication removal for samples with different microbial contents with co-preparation and DNA-preparation.The simulated microbial community samples were prepared using the NadPrep RNA & DNA Library Co-Preparation Module (Co-Prep) and NadPrep EZ DNA Library
Preparation Module v2 (DNA-Prep), in conjunction with NadPrep Universal Stubby Adapter (UDI) Adapter for library preparation, followed by hybrid capture using
NadPrep ES Hybrid Capture Reagents and the NEX-t Panel v1.0.
Note: Simulated microbial community samples of 0.001% - 1% MSA-1003 were created by diluting a mixture of 20 strains of genomic material (ATCC, MSA-1003) using the human genomic DNA standard (Promega, G1471) at various ratios. Sequencing platform: Illumina NovaSeq 6000, PE150.
Low GC Bias
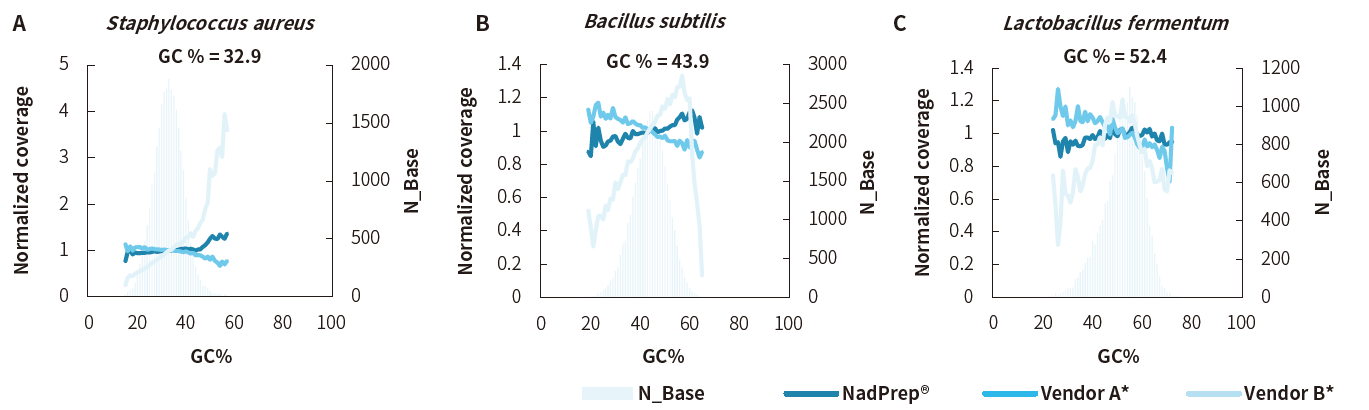
Figure 6. Uniform coverage of regions with various GC content using NadPrep RNA & DNA Library Co-Preparation Module. A. Staphylococcus aureus, B. Bacillus subtilis, and C. Lactobacillus fermentum with different GC content.
Application Example in Clinical Sample
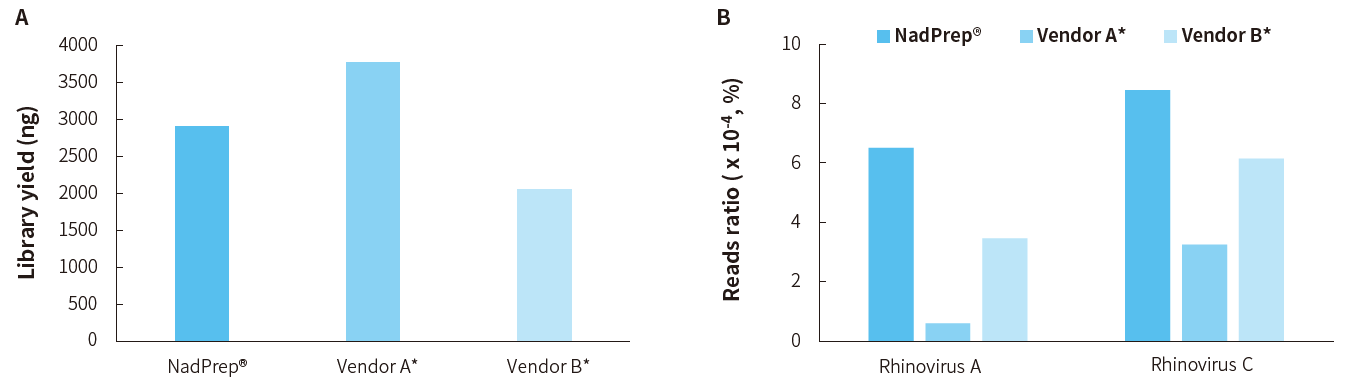
Figure 7. Detection performance of tNGS Total Solution for clinical samples with NadPrep RNA & DNA co-preparation. A. Library yield; B. Reads ratio of
Rhinovirus A and Rhinovirus C.
Note: The samples were derived from 50 ng of clinical rhinovirus nucleic acid samples. Sequencing platform: Illumina NovaSeq 6000, PE150
Library Module
| Package | Color of Cap Tube | Component |
Volume 1002411(24 rxn) |
Volume 1002412 (96 rxn) |
| Box1 |
|
Co-Random Primer | 60 µL | 230 µL |
|
|
1st Co-Strand Synth. Buffer | 120 µL | 460 µL | |
|
|
1st Co-strand Synth.Enzyme | 60 µL | 230 µL | |
|
|
2nd Co-strand Synth.Buffer | 240 µL | 960 µL | |
|
|
2nd Co-strand Synth. Enzyme | 60 µL | 230 µL | |
|
|
Co-FERA Buffer | 150 µL | 575 µL | |
|
|
Co-FERA Enzyme |
150 μL |
575 μL |
|
|
|
Ligation Buffer | 750 μL | 2×1500 μL | |
|
|
DNA Ligase | 65 μL | 250 µL | |
|
|
2X HiFi PCR Master Mix | 720 μL | 2×1600 µL | |
|
/ |
Nuclease Free Water | 2.5 mL | 8 mL | |
|
|
TE Solution | 1 mL | 4 mL | |
| Box2 |
/ |
NadPrep SP Beads | 4 mL | 15 mL |
Universal Adapter (MDI) Module (for MGI)
| Color of Cap Tube | Component |
Volume 1003711 (24 rxn) |
Volume 1003721 (96 rxn) |
Volume 1003722 (96 rxn) |
|
|
NadPrep M-Index (MDI) Primer Mix # | 12×15 µL (Tube) | 24×25 µL (Tube) | 24×25 µL (Tube) |
|
|
NadPrep M-Adapter (DI) | 60 µL (Tube) | 230 µL (Tube) | 230 µL (Tube) |
| Color of Cap Tube | Component |
Volume 1003751 (96 rxn) |
Volume 1003752 (96 rxn) |
Volume 1003753 (96 rxn) |
Volume 1003754 (96 rxn) |
|
|
NadPrep M-Index (MDI) Primer Mix # | 96×8 µL (Plate) | 96×8 µL (Plate) | 96×8 µL (Plate) | 96×8 µL (Plate) |
|
|
NadPrep M-Adapter (DI) | 260 µL (Tube) | 260 µL (Tube) | 260 µL (Tube) | 260 µL (Tube) |
|
|
NadPrep M-Amplification Primer Mix (for MGI, DI) | 75 µL (Tube) | 75 µL (Tube) | 75 µL (Tube) | 75 µL (Tube) |
Universal Stubby Adapter (UDI) Module
| Color of Cap Tube | Component |
Volume 1003240 (24 rxn) |
Volume 1003241 (96 rxn) |
Volume 1003242 (96 rxn) |
|
|
NadPrep Universal UDI-index Primer Mix # | 12×15 µL (Tube) | 24×25 µL (Tube) | 24×25 µL (Tube) |
|
|
NadPrep Universal Stubby Adapter | 60 µL (Tube) | 230 µL (Tube) | 230 µL (Tube) |
|
|
NadPrep Amplification Primer Mix II | 20 µL (Tube) | 75 µL (Tube) | 75 µL (Tube) |
| Color of Cap Tube | Component |
Volume 1003251 (96 rxn) |
Volume
1003252 (96 rxn) |
Volume
1003253 (96 rxn) |
Volume
1003254 (96 rxn) |
|
|
NadPrep Universal UDI-index Primer Mix # | 96×8 µL (Plate) | 96×8 µL (Plate) | 96×8 µL (Plate) | 96×8 µL (Plate) |
|
|
NadPrep Universal Stubby Adapter | 260 µL (Tube) | 260 µL (Tube) | 260 µL (Tube) | 260 µL (Tube) |
|
|
NadPrep Amplification Primer Mix II | 75 µL (Tube) | 75 µL (Tube) | 75 µL (Tube) | 75 µL (Tube) |
√ If the sample is a co-extracted total nucleic acid, quantification of RNA and DNA should be performed separately using Qubit, and library amplification should be carried out according to the recommended cycles in the manual.
√ During the experiment, RNase-free consumables must be used, and the operating environment should be free of RNase contamination, as this can affect the efficiency of RNA reverse transcription.
√ If the nucleic acid sample contains excessive amounts of chelating agents, guanidine salts, phenol, proteins, ethanol, or other impurities, it can impact the activity of RNA reverse transcriptase and the efficiency of DNA enzymatic digestion. It is recommended refer to purify the nucleic acid samples using 2X NadPrep SP Beads, replace the elution reagent with Nuclease Free Water, and then proceed with library preparation.
√ For samples of lower quality, consider increasing the number of PCR amplification cycles appropriately.
If the nucleic acid samples contain high concentrations of EDTA or have an excessively high pH, it can affect the efficiency of DNA digestion. It is recommended to purify nucleic acid samples using 2X NadPrep SP Beads, replacing the elution reagent with Nuclease Free Water before proceeding with library preparation.
This module can tolerate nucleic acid samples containing 6 μL of EDTA (1 mM), resulting in a final concentration of 0.3 mM in the cDNA first-strand synthesis system. At this concentration, RNA reverse transcription inhibition is minimal, and the efficiency of enzymatic digestion is unaffected. Before library preparation, confirm the sample solvent. If it exceeds the recommended EDTA threshold, it is necessary to purify nucleic acid samples using 2X NadPrep SP Beads, replacing the elution reagent with Nuclease Free Water before proceeding with library preparation.
When co-prepare RNA & DNA libraries from mixed nucleic acid samples, a higher proportion of DNA samples results in larger fragments with enzymatic digestion. Adjustments can be made according to the expected insert fragment lengths and recommended time of enzymatic fragmentation in the manual.
Libraries prepared with this module can be used in conjunction with NEX-t Panel v1.0 and NadPrep ES Hybrid Capture Reagents. This significantly shortens the time required for pathogen-targeted sequencing and simplifies experimental procedures.
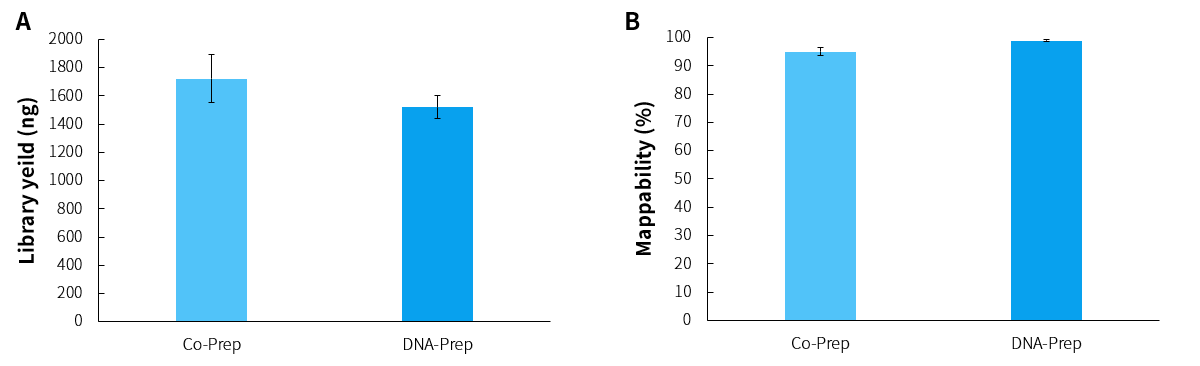
Figure 1. Library yield and capture performance of DNA samples using different library preparation protocols. A. Library Yield; B. Mappability.
Note: The sample originated from 50 ng of Human Genomic DNA Standard (Promega, G1521). Sequencing mode was Illumina NovaSeq 6000 platform, PE150.
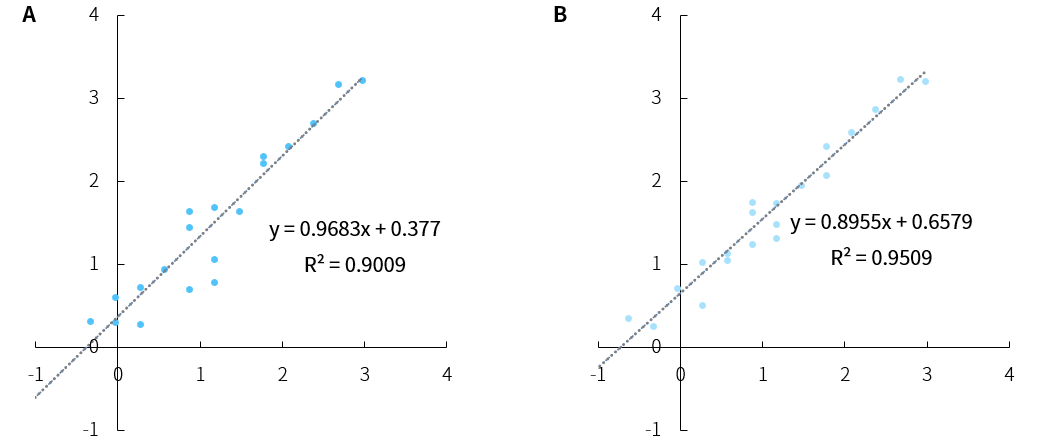
Figure 2. Correlation of ERCC expression profiles in captured libraries from RNA samples processed through different library preparation protocols. A. NadPrep RNA & DNA Library Co-Preparation Module; B. NadPrep Total RNA-To-DNA Module conjunction NadPrep DNA Library Preparation Module.
Note: The samples originated from K562 cell line RNA with an initial input of 50 ng; the pre-library input amount was 200 ng.
| Type | Product | Detail | Catalog# |
| Library Module | NadPrep RNA & DNA Library Co-Preparation Module, 24 rxn | 24 rxn | 1002411 |
| NadPrep RNA & DNA Library Co-Preparation Module, 96 rxn | 96 rxn | 1002412 | |
| Universal Adapter (MDI) Module | NadPrep Universal Adapter (MDI) Module Set A1 (for MGI), 24 rxn |
MDI 1-12 24 rxn |
1003711 |
| NadPrep Universal Adapter (MDI) Module Set B1 (for MGI), 96 rxn |
MDI 1-24 96 rxn |
1003721 | |
| NadPrep Universal Adapter (MDI) Module Set B2 (for MGI), 96 rxn |
MDI 25-48 96 rxn |
1003722 | |
| NadPrep Universal Adapter (MDI) Module Set E1 (for MGI), 96 rxn |
MDI 1-96 96 rxn |
1003751 | |
| NadPrep Universal Adapter (MDI) Module Set E2 (for MGI), 96 rxn |
MDI 97-192 96 rxn |
1003752 | |
| NadPrep Universal Adapter (MDI) Module Set E3 (for MGI), 96 rxn |
MDI 193-288 96 rxn |
1003753 | |
| NadPrep Universal Adapter (MDI) Module Set E4 (for MGI), 96 rxn |
MDI 289-384 96 rxn |
1003754 | |
| Universal Stubby Adapter (UDI) Module | NadPrep Universal Stubby Adapter (UDI) Module Set A1, 24 rxn |
UDI 1-12 24 rxn |
1003240 |
| NadPrep Universal Stubby Adapter (UDI) Module Set B1, 96 rxn |
UDI 1-24 96 rxn |
1003241 | |
| NadPrep Universal Stubby Adapter (UDI) Module Set B2, 96 rxn |
UDI 25-48 96 rxn |
1003242 | |
| NadPrep Universal Stubby Adapter (UDI) Module Set E1, 96 rxn |
UDI 1-96 96 rxn |
1003251 | |
| NadPrep Universal Stubby Adapter (UDI) Module Set E2, 96 rxn |
UDI 97-192 96 rxn |
1003252 | |
| NadPrep Universal Stubby Adapter (UDI) Module Set E3, 96 rxn |
UDI 193-288 96 rxn |
1003253 | |
| NadPrep Universal Stubby Adapter (UDI) Module Set E4, 96 rxn |
UDI 289-384 96 rxn |
1003254 |
For research use only. Not for use in diagnostic procedures.
Product Sheet
-
[Product Catalog] -
Product Catalog
Download
-
[Technical Note] -
Checklist_EN_NadPrep RNA & DNA Library Co-Preparation Module
Download
-
[Poster] -
Brochure-NadPrep RNA & DNA Library Co-Preparation Module
Download
-
[Technical Note] -
EN_Protocol_NadPrep_RNA & DNA_Library_Co-Preparation_Module_V1.0
Download
-
[MSDS] -
NadPrep RNA & DNA Library Co-Preparation Module, 96 rxn
Download
-
[MSDS] -
NadPrep RNA & DNA Library Co-Preparation Module, 24 rxn
Download
Application Note
Demo Data
Solutions
- Methyl Library Preparation Total Solution
- Sequencing single library on different platform--Universal Stubby Adapter (UDI)
- HRD score Analysis
- Unique Dual Index for MGI platforms
- RNA-Cap Sequencing of Human Respiratory Viruses Including SARS-CoV-2
- Total Solution for RNA-Cap Sequencing
- Total Solution for MGI Platforms
- Whole Exome Sequencing
- Low-frequency Mutation Analysis
Events
-
Exhibition Preview | Nanodigmbio invites you to join us at Boston 2025 Annual Meeting of the American Society of Human Genetics (ASHG)

-
Exhibition Preview | Nanodigmbio Invites You to Join Us at WHX & WHX Labs Kuala Lumpur 2025, Malaysia International Trade and Exhibition Centre in Kuala Lumpur

-
Exhibition Preview | Nanodigmbio Invites You to Join Us at Hospitalar 2025, Brazil International Medical Device Exhibition in São Paulo

-
Exhibition Preview | Nanodigmbio invites you to join us at Denver 2024 Annual Meeting of the American Society of Human Genetics (ASHG)

-
Exhibition Preview | Nanodigmbio invites you to join us at Sapporo 2024 Annual Meeting of the Japan Society of Human Genetics (JSHG)

-
Exhibition Preview | Nanodigmbio invites you to join us at Association for Diagnostics & Laboratory Medicine (ADLM)

Nanodigmbio will contact you within 1 day.
If there are any problems, please contact us by 400 8717 699 / support@njnad.com



