
NanoHema Panel v2.0
#1001721
NanoHema
Panel v2.0 targets 481 genes
associated with hematologic malignancies, comprehensively covering a wide range
of genetic alterations including base substitutions, insertions/deletions, amplifications,
and gene fusions.
This
panel consists of two components:
NanoHema
Panel v2.0_DNA Tube: Covers 1.7 Mb of genomic regions, including full coding regions of 436 genes,
partial intronic regions of 12 genes, and the IGH Switch regions.
NanoHema
Panel v2.0_RNA Tube: Covers
146 genes, optimized for fusion gene detection.
By integrating multi-omics approaches, employing a highly targeted design, and enhancing clinical utility, NanoHema Panel v2.0 offers an efficient and cost-effective solution for precision diagnosis, treatment planning, and longitudinal management of hematologic malignancies.
- Product details
- Kit Content
- FAQ
- Ordering Information
- Resources
Feature
● Comprehensive Multi-type Mutation Coverage
Encompasses 481 hematologic malignancy-related genes, enabling thorough analysis of base substitutions, indels, amplifications, and gene fusions.
● Dual Workflow for Enhanced Accuracy
RNA workflow compensates for the limitations of the DNA workflow by accurately identifying fusion gene partners and validating the functional impact of fusion events. The synergistic use of both workflows ensures comprehensive and precise detection.
● Flexible Customization
Supports flexible subdivision into multiple sub-panels or the addition of target genes, allowing adaptation to diverse hematologic malignancy diagnostic needs in both research and clinical applications.
Performance
Capture Performance_NanoHema Panel v2.0_DNA
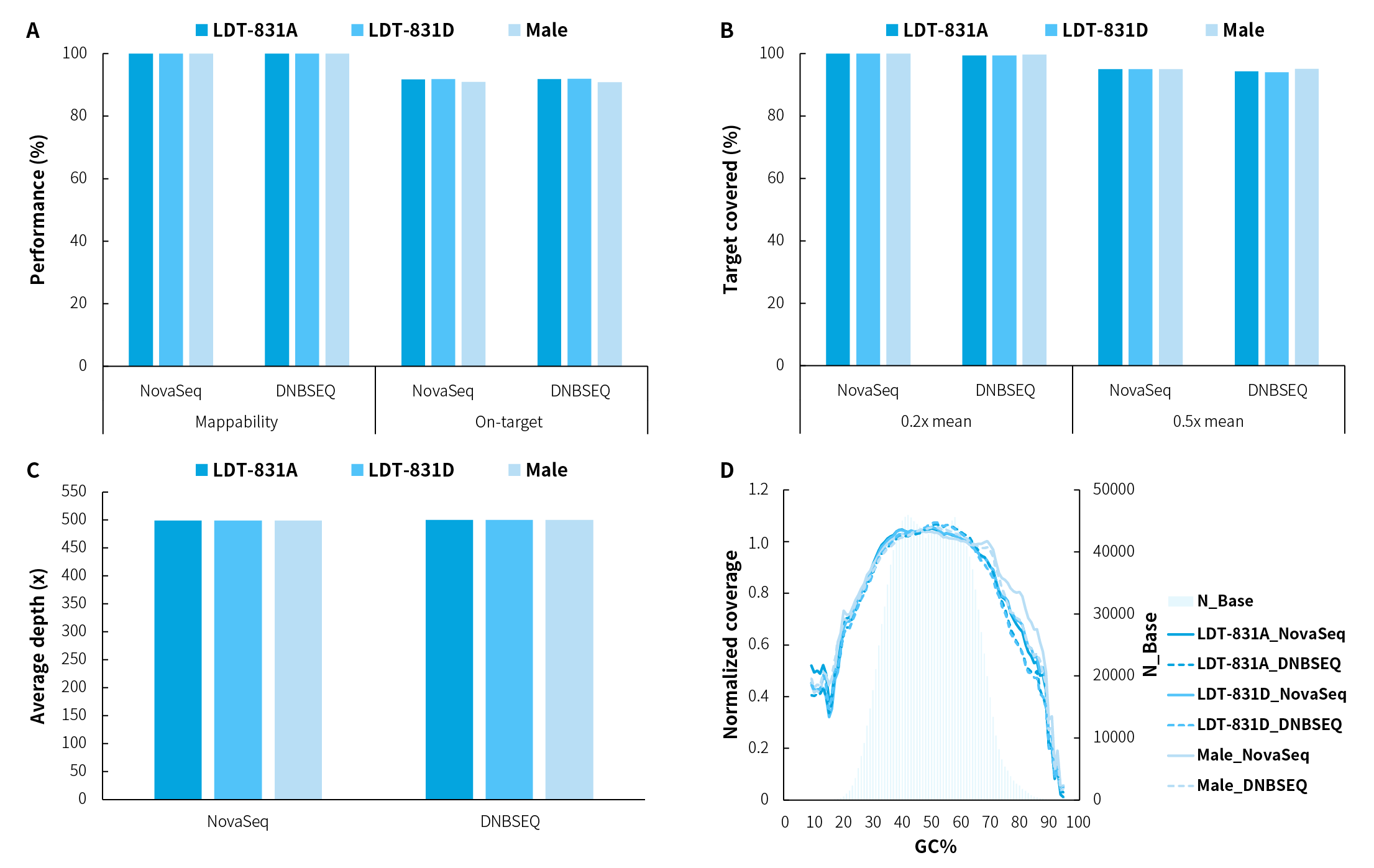
Figure 1. Capture performance of NanoHema Panel v2.0_DNA applied to various types of gDNA samples. A. Mappability & On-target rate; B. Target covered; C. Average sequencing depth after deduplication; D. GC bias. Using 50 ng of various gDNA samples, pre-libraries were prepared with the NadPrep EZ DNA Library Preparation Kit coupled with the NadPrep Universal Stubby Adapter (UDI) Module, followed by hybrid capture using NanoHema Panel v2.0_DNA with NadPrep Hybrid Capture Reagents. Sequencing was performed on both NovaSeq 6000 (PE150) and DNBSEQ-T7 (PE150) platforms, with a random subset of 1.3 Gb used for data analysis.
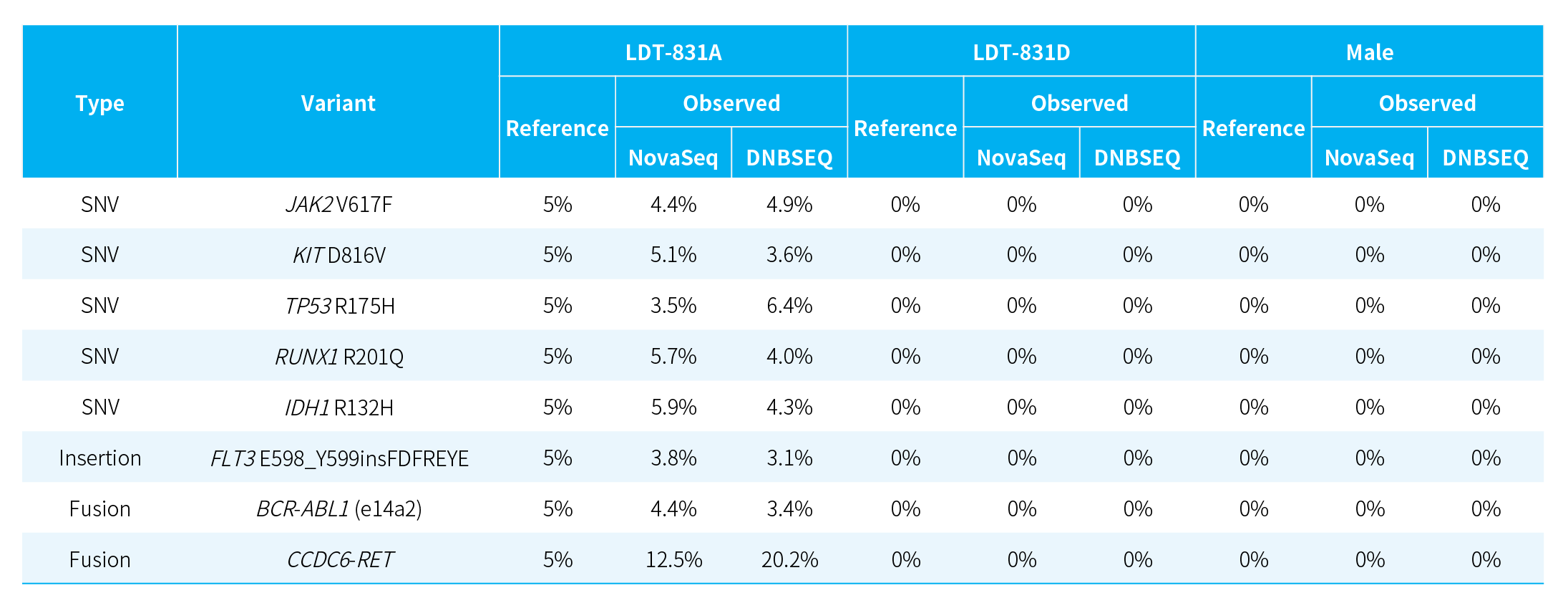
Note: LDT-831: hematologic malignancy gDNA quality control (LDT Bioscience; LDT-831A: AF = 5%; LDT-831D: AF = 0%). Male: human genomic DNA standard (Promega, G1471).
Stable Detection of Multiple Mutation Types_NanoHema Panel v2.0_DNA
Table 1. Detection of various mutation types in different standard samples using NanoHema Panel v2.0_DNA.
Capture Performance_NanoHema Panel v2.0_RNA
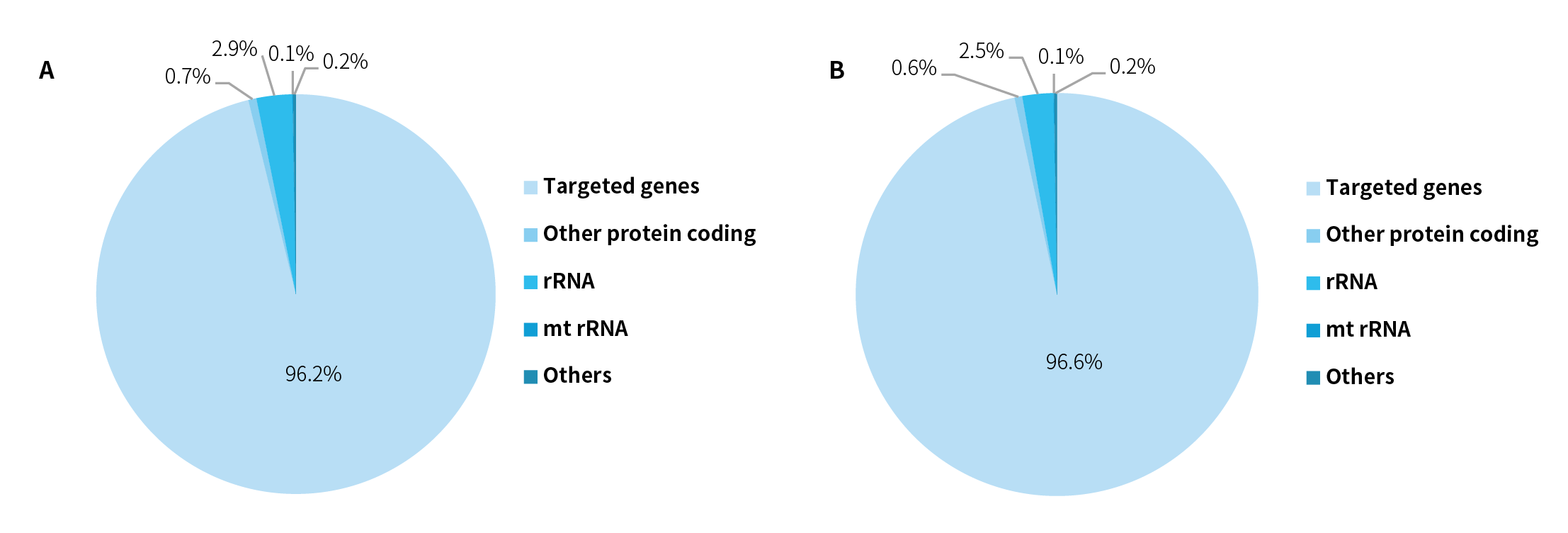
Figure 2. Composition of capture sequencing data using NanoHema Panel v2.0_RNA. For the K562 cell line RNA (50 ng), pre-libraries were prepared using the NadPrep Total RNA-To-DNA Module coupled with the NadPrep DNA Library Preparation Kit v2, followed by hybrid capture with NanoHema Panel v2.0_RNA and NadPrep Hybrid Capture Reagents. Sequencing was performed on NovaSeq 6000 (PE150) (A) and DNBSEQ-T7 (PE150) (B), with analysis performed using Salmon.
Fusion Detection_NanoHema Panel v2.0_RNA
Table 2. Detection of fusion genes in K562 cell line RNA using NanoHema Panel v2.0_RNA.
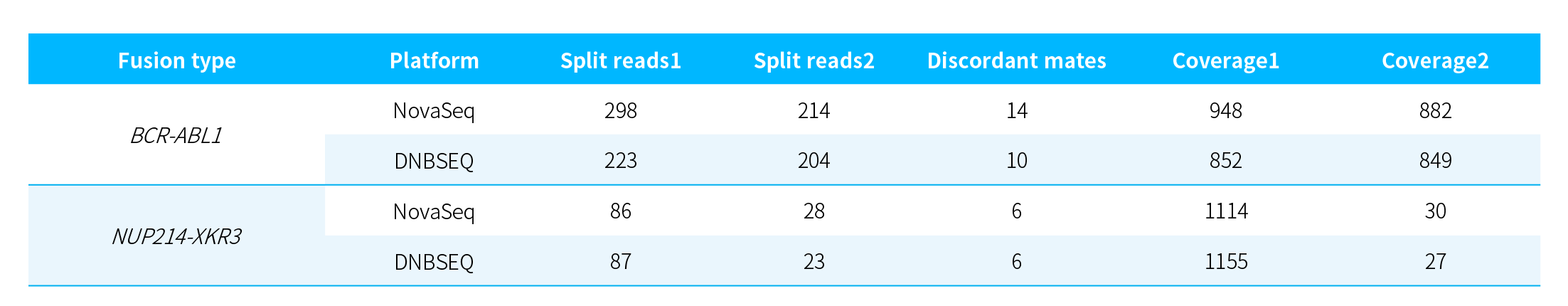
Note: Data are reported per M read pairs (approximately 0.3 Gb); analysis was performed using Arriba.
Gene List
|
NanoHema Panel v2.0_DNA_436 genes_Full CDS |
||||||||||||||
| ABL1 | ABL2 | ACTB | AKT1 | AKT2 | AKT3 | ALK | AMER1 | ANKRD26 | AP3B1 | APC | AR | ARAF | ARHGAP26 | ARID1A |
| ARID1B | ARID2 | ARID5B | ASXL1 | ASXL2 | ASXL3 | ATG5 | ATM | ATR | ATRX | ATXN1 | AURKA | AURKB | AXIN1 | AXL |
| B2M | BAALC | BAP1 | BARD1 | BCL10 | BCL11A | BCL11B | BCL2 | BCL6 | BCL7A | BCOR | BCORL1 | BIRC3 | BLM | BLNK |
| BRAF | BRCA1 | BRCA2 | BRD4 | BTG1 | BTG2 | BTK | BTLA | CALR | CARD11 | CBFB | CBL | CBLB | CCND1 | CCND2 |
| CCND3 | CCNE1 | CCR4 | CCR7 | CCT6B | CD22 | CD27 | CD274 | CD28 | CD58 | CD70 | CD79A | CD79B | CD83 | CDC42 |
| CDC73 | CDH1 | CDK12 | CDK4 | CDK6 | CDK8 | CDKN1A | CDKN1B | CDKN1C | CDKN2A | CDKN2B | CDKN2C | CEBPA | CEP57 | CHD2 |
| CHD8 | CHEK1 | CHEK2 | CIITA | CKS1B | CREBBP | CRLF2 | CSF1R | CSF3R | CTCF | CTLA4 | CTNNB1 | CUX1 | CXCR4 | CYLD |
| DAXX | DDR2 | DDX3X | DDX41 | DHX15 | DNM2 | DNMT3A | DOT1L | DTX1 | DUSP2 | DUSP22 | EBF1 | ECT2L | EED | EGFR |
| EP300 | EPCAM | EPHA2 | EPHA3 | EPHA5 | EPHA7 | EPOR | ERBB2 | ERBB3 | ERBB4 | ERCC4 | ERG | ESR1 | ETNK1 | ETS1 |
| ETV1 | ETV4 | ETV6 | EZH2 | FAM46C | FANCA | FANCC | FANCD2 | FANCE | FANCF | FANCG | FANCI | FANCJ | FANCL | FANCM |
| FAS | FAT1 | FAT4 | FBXO11 | FBXW7 | FGFR1 | FGFR2 | FGFR3 | FGFR4 | FH | FLCN | FLT1 | FLT3 | FLT4 | FOXO1 |
| FOXO3 | FOXP1 | FYN | GADD45B | GATA1 | GATA2 | GATA3 | GNA11 | GNA13 | GNAQ | GNAS | GNB1 | GRIN2A | HACE1 | HBA1 |
| HBA2 | HBB | HDAC1 | HDAC4 | HDAC7 | HGF | HIST1H1A | HIST1H1C | HIST1H1D | HIST1H1E | HNF1A | HRAS | ID3 | IDH1 | IDH2 |
| IFNAR2 | IGF1R | IGH | IGK | IGL | IGLL5 | IKBKE | IKZF1 | IKZF2 | IKZF3 | IL3 | IL7R | INPP4B | INPP5D | IRF1 |
| IRF4 | IRF8 | ITK | ITPKB | JAK1 | JAK2 | JAK3 | JARID2 | JUN | KAT6A | KDM2B | KDM5A | KDM5C | KDM6A | KDR |
| KIT | KLF2 | KLHL5 | KLHL6 | KMT2A | KMT2B | KMT2C | KMT2D | KRAS | LEF1 | LMO2 | LRP1B | LTB | LYN | LYST |
| MAF | MAFB | MALT1 | MAP2K1 | MAP2K2 | MAP2K4 | MAP3K1 | MAP3K14 | MAPK1 | MCL1 | MDM2 | MDM4 | MED12 | MEF2B | MEF2D |
| MEN1 | MET | MFHAS1 | MGA | MITF | MLH1 | MPL | MSH2 | MSH3 | MSH6 | MTOR | MUTYH | MYB | MYC | MYCL |
| MYCN | MYD88 | NBN | NCKAP1L | NCSTN | NF1 | NF2 | NFE2L2 | NFKB1 | NFKB2 | NFKBIA | NFKBIE | NKX2-1 | NLRC4 | NOTCH1 |
| NOTCH2 | NPM1 | NRAS | NSD1 | NT5C2 | NTRK1 | NTRK3 | NUP98 | P2RY8 | PAG1 | PAK3 | PALB2 | PAX5 | PBRM1 | PC |
| PDCD1 | PDCD1LG2 | PDGFRA | PDGFRB | PDK1 | PHF6 | PIK3CA | PIK3CD | PIK3R1 | PIK3R2 | PIM1 | PLCG1 | PLCG2 | PML | PMS2 |
| POLE | POT1 | POU2AF1 | PPM1D | PPP2R1A | PRDM1 | PRF1 | PRKCB | PRPF8 | PTCH1 | PTEN | PTPN1 | PTPN11 | PTPN2 | PTPN6 |
| PTPRD | PTPRK | PTPRO | RAB27A | RAD21 | RAD50 | RAD51 | RAF1 | RARA | RASGEF1A | RB1 | RECQL4 | REL | RELN | RET |
| RHOA | RICTOR | RNF43 | ROS1 | RPL10 | RPS15 | RPTOR | RUNX1 | SAMD9 | SAMD9L | SBDS | SDHA | SDHB | SDHC | SDHD |
| SETBP1 | SETD2 | SF3B1 | SGK1 | SH2B3 | SH2D1A | SLC19A1 | SLC7A7 | SMAD2 | SMAD4 | SMARCA2 | SMARCA4 | SMARCB1 | SMC1A | SMC3 |
| SMO | SOCS1 | SOX11 | SOX2 | SPEN | SPOP | SRC | SRP72 | SRSF2 | STAG2 | STAT3 | STAT4 | STAT5A | STAT5B | STAT6 |
| STK11 | STX11 | STXBP2 | SUFU | SUZ12 | SYK | TBL1XR1 | TCF3 | TCL1A | TERC | TERT | TET1 | TET2 | TET3 | TGFBR2 |
| TNFAIP3 | TNFRSF11A | TNFRSF14 | TNFRSF17 | TNFRSF1B | TOP1 | TOP2A | TP53 | TP63 | TP73 | TRAF2 | TRAF3 | TRAF5 | TSC1 | TSC2 |
| TSHR | TYK2 | U2AF1 | UNC13D | VAV1 | VHL | WHSC1 | WT1 | XIAP | XPC | XPO1 | ZAP70 | ZBTB7A | ZEB2 | ZMYM3 |
| ZRSR2 | ||||||||||||||
| NanoHema Panel v2.0_DNA_12 genes_Selected Intron | ||||||||||||||
| ALK | BCL6 | BCR | BRAF | ETV6 | JAK2 | KMT2A | MYC | PDGFRA | PDGFRB | RARA | RET | |||
| NanoHema Panel v2.0_RNA_146 genes | ||||||||||||||
| ABL1 | ABL2 | ALK | ARHGAP26 | ARID1A | ASXL1 | ATG5 | BCL10 | BCL11A | BCL11B | BCL2 | BCL3 | BCL6 | BCL7A | BCL9 |
| BCOR | BCR | BIRC3 | BRAF | BTG1 | CBFA2T3 | CBFB | CBL | CCND1 | CCND2 | CCND3 | CD274 | CDK6 | CEBPE | CIITA |
| CREBBP | CRLF2 | CTNNB1 | DEK | DUSP22 | DUX4 | EGFR | EP300 | EPOR | ERBB2 | ERG | ETS1 | ETV1 | ETV4 | ETV6 |
| EWSR1 | FGFR1 | FGFR2 | FGFR3 | FOXO1 | FOXO3 | FOXP1 | GLIS2 | HLF | IGH | IGK | IGL | IKZF1 | IL3 | IRF4 |
| ITK | JAK1 | JAK2 | JAK3 | KAT6A | KIF5B | KMT2A | LMO1 | LMO2 | LYN | MAF | MAFB | MALT1 | MECOM | MEF2D |
| MLLT1 | MLLT10 | MLLT3 | MN1 | MTCP1 | MYB | MYC | MYH11 | NCOA2 | NCOR1 | NF1 | NF2 | NFKB2 | NOTCH1 | NPM1 |
| NSD1 | NTRK1 | NTRK3 | NUP214 | NUP98 | NUTM1 | P2RY8 | PAX5 | PBX1 | PDCD1LG2 | PDGFRA | PDGFRB | PICALM | PIM1 | PML |
| POU2AF1 | PRDM1 | PTCH1 | RAF1 | RARA | RARB | RARG | RBM15 | RET | ROS1 | RUNX1 | RUNX1T1 | STAT6 | STIL | SYK |
| TAF15 | TAL1 | TBL1XR1 | TCF3 | TCL1A | TCL1B | TET1 | TLX1 | TLX3 | TNFRSF11A | TOP1 | TP63 | TPM3 | TPM4 | TRA |
| TRB | TRD | TRG | TYK2 | VAV1 | WHSC1 | ZBTB16 | ZEB2 | ZNF292 | ZNF362 | ZNF384 | ||||
| #Package | Cap Color | Component |
Volume 1001721 (96 rxn) |
Volume 1001722 (16 rxn) |
| Box 1 |
|
NanoHema Panel v2.0_DNA | 415 μL | 70 μL |
|
|
NanoHema Panel v2.0_RNA | 415 μL | 70 μL |
Fusion
genes serve as critical molecular markers for the diagnosis and treatment of
hematologic malignancies, making their accurate detection essential. The DNA
workflow detects gene fusions by designing probes covering intronic regions;
however, due to the large size and high repetitiveness of introns, relying
solely on DNA detection poses challenges such as high cost and incomplete
coverage, and it cannot confirm the functional impact of fusion events. In
contrast, the RNA workflow directly focuses on the transcript level, enabling
precise identification of actual fusion events and their partner genes while
assessing whether these fusions lead to aberrant transcription. Consequently,
the combined use of DNA and RNA workflows provides a “dual safeguard” for fusion gene detection, enhancing both the
comprehensiveness and accuracy of the results.
Total
RNA-seq requires rRNA depletion and individual reactions per sample, increasing
both complexity and cost. Moreover, total RNA-seq exhibits relatively lower
sensitivity, particularly in detecting low-frequency fusion events in
low-expression samples. In contrast, the DNA panel in NanoHema Panel v2.0
already covers the intronic regions of key genes, allowing users to flexibly
add custom intronic regions based on requirements and budget without the need
for a separate RNA panel.
NanoHema Panel v2.0 is a comprehensive large panel that covers a wide range of hematologic malignancy-associated gene variants. To meet diverse clinical needs, the panel offers multiple customization options:
• It can be flexibly split into multiple sub-panels (e.g., for acute myeloid leukemia [AML], lymphoblastic leukemia, T-cell lymphoma, and B-cell lymphoma).
• Additional target genes can be incorporated based on specific research or diagnostic requirements, enabling the creation of a personalized detection solution.
| Product | Catalog# |
| NanoHema Panel v2.0, 96 rxn | 1001721 |
| NanoHema Panel v2.0, 16 rxn | 1001722 |
Product Sheet
-
[Product Catalog] -
Product Catalog
Download
-
[Poster] -
EN-Brochure-NanoHema Panel v2.0
Download
-
[Technical Note] -
EN-Instruction-NanoHema Panel v2.0-Version 1.0
Download
-
[Bed File] -
NanoHema Panel v2.0
Download
-
[Technical Note] -
Protocol-NadPrep ES Hybrid Capture of DNA Libraries (for Illumina) v1.1
Download
-
[Technical Note] -
Protocol_NadPrep ES Hybrid Capture of DNA Libraries (for MGI) v1.1
Download
-
[Technical Note] -
Hybridization Capture of DNA Libraries (for MGI) Protocol v1.6
Download
-
[Technical Note] -
Hybridization Capture of DNA Libraries (for for Illumina®) Protocol v2.5
Download
Application Note
Demo Data
Solutions
- Methyl Library Preparation Total Solution
- Sequencing single library on different platform--Universal Stubby Adapter (UDI)
- HRD score Analysis
- Unique Dual Index for MGI platforms
- RNA-Cap Sequencing of Human Respiratory Viruses Including SARS-CoV-2
- Total Solution for RNA-Cap Sequencing
- Total Solution for MGI Platforms
- Whole Exome Sequencing
- Low-frequency Mutation Analysis
Events
-
Exhibition Preview | Nanodigmbio invites you to join us at Boston 2025 Annual Meeting of the American Society of Human Genetics (ASHG)

-
Exhibition Preview | Nanodigmbio Invites You to Join Us at WHX & WHX Labs Kuala Lumpur 2025, Malaysia International Trade and Exhibition Centre in Kuala Lumpur

-
Exhibition Preview | Nanodigmbio Invites You to Join Us at Hospitalar 2025, Brazil International Medical Device Exhibition in São Paulo

-
Exhibition Preview | Nanodigmbio invites you to join us at Denver 2024 Annual Meeting of the American Society of Human Genetics (ASHG)

-
Exhibition Preview | Nanodigmbio invites you to join us at Sapporo 2024 Annual Meeting of the Japan Society of Human Genetics (JSHG)

-
Exhibition Preview | Nanodigmbio invites you to join us at Association for Diagnostics & Laboratory Medicine (ADLM)

Nanodigmbio will contact you within 1 day.
If there are any problems, please contact us by 400 8717 699 / support@njnad.com


