New Arrival | Upgraded Library Preparation Kit, Easily Achieve Higher Yield and Conversion Rate!
View: 5256 / Time: 2024-08-29
01 Background
DNA library preparation is the first step in any Next Generation Sequencing (NGS) workflow and is the process whereby DNA or RNA are processed prior to sequencing. It begins with fragmentation, either mechanical or enzymatic to shear the genomic DNA from the target samples into the appropriate insert size. Subsequently, the ends of the DNA are then repaired and primed for the ligation of specific sequencing adapters which are added to the 3’ and 5’ ends. This process results in the formation of an effective library that meets the requirements of the sequencing platform.
The entire process of a routine NGS experiment includes nucleic acid extraction, library preparation, target capture, sequencing, and data analysis. Among them, library preparation, as a core step in the wet-lab NGS workflow, is crucial for the successful conduct of the NGS process. Therefore, the quality of the library prepared using different library preparation kits directly affects the accuracy and reliability of the final sequencing results. With the continuous advancement of NGS technology, NGS has been widely applied in various fields such as basic research, genetic disease detection, non-invasive prenatal testing (NIPT), tumor companion diagnostics (CDx), pre-implantation genetic screening (PGS), and identification of infectious pathogens. Users have placed higher demands for the stability, effectiveness, accuracy, and efficiency of sequencing libraries. From this perspective, it is particularly important to improve the convenience of the NGS library preparation process and the conversion rate (the proportion of initial DNA successfully converted into a sequencable library).
Therefore, in order to meet market demands, Nanodigmbio has launched an upgraded library preparation kit: NadPrep DNA Library Preparation Module v2, which can complete whole-genome library preparation within 3 hours. Compared to the v1 version, the v2 version has improved the library yield and better tolerance for low-quality samples, as well as increased the library conversion rate. Additionally, NadPrep DNA Library Preparation Module v2 is compatible with various types of adapters for different sequencing platforms already launched by Nanodigmbio, allowing for flexible and free combinations.
02 Upgraded Version Library Preparation Kit
2.1 Introduction
NadPrep DNA Library Preparation Module v2 is designed for the preparation of high-quality libraries from double-stranded DNA (dsDNA) on Illumina® & MGI platforms. This A-T ligation-based kit offers a stable and efficient library preparation solution for applications including whole genome sequencing of 1‒500 ng DNA and hybridization capture based targeted sequencing. Compared to the v1 version, the upgraded kit has improved the library conversion efficiency and better tolerance for low-quality samples. Consequently, it can enhance library complexity while maintaining high fidelity.
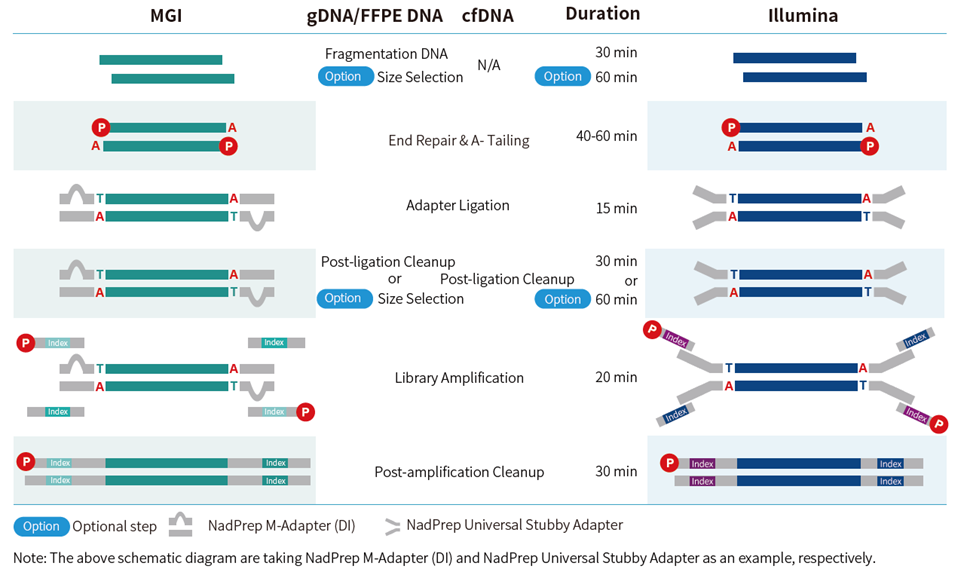
Figure 1. Entire experimental process of library preparation with NadPrep DNA Library Preparation Module v2.
2.2 Features
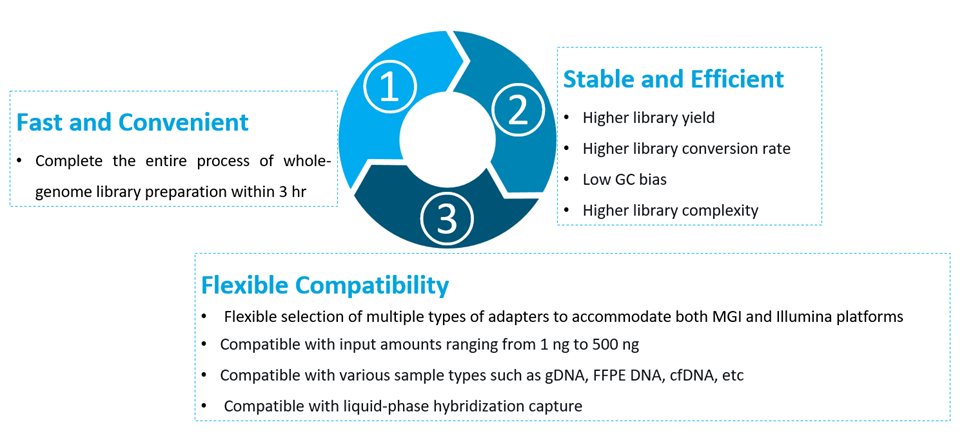
Figure 2. Features of NadPrep DNA Library Preparation Module v2.
03 Performance
3.1 Stable and Efficient Library Yield
3.1.1 Compatible with a wide range of input amounts
Using the human genomic DNA standard (Promega, G1521) as a template with different initial input amounts, library preparation was performed using NadPrep DNA Library Preparation Module v2. The results indicate that the library yield at each initial input amount meets the expected output, demonstrating the suitability of this kit for sample initial input amounts ranging from 1 ng to 500 ng.
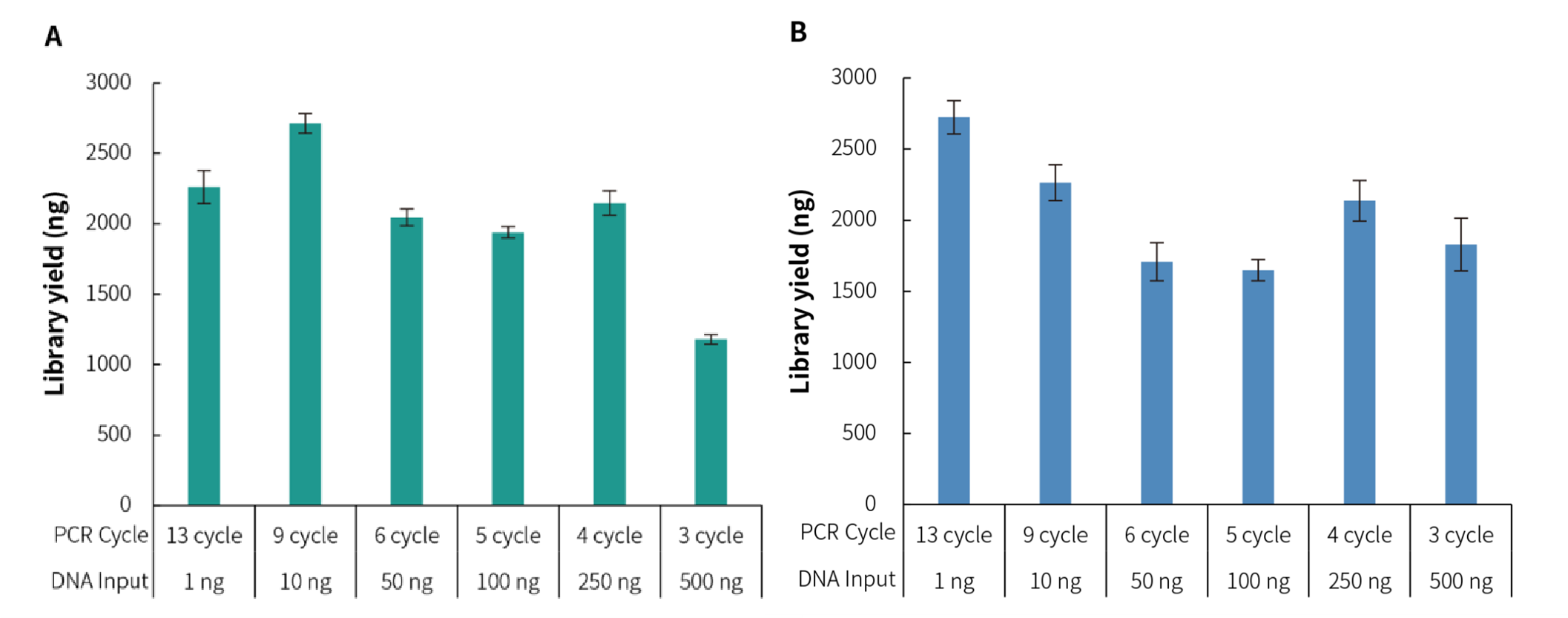 Figure 3. Library yield of NadPrep DNA Library Preparation Kit v2 for gDNA samples with different input amounts. The libraries were prepared by using NadPrep DNA Library Preparation Module v2 coupled with NadPrep Universal Adapter (MDI) Module (for MGI) and NadPrep Universal Stubby Adapter (UDI) Module. A. Library yield (MGI); B. Library yield (Illumina).
Figure 3. Library yield of NadPrep DNA Library Preparation Kit v2 for gDNA samples with different input amounts. The libraries were prepared by using NadPrep DNA Library Preparation Module v2 coupled with NadPrep Universal Adapter (MDI) Module (for MGI) and NadPrep Universal Stubby Adapter (UDI) Module. A. Library yield (MGI); B. Library yield (Illumina).
Note: gDNA samples are human genomic DNA standard (Promega, G1471).
3.1.2 Compatible with various types of samples
For different types of samples, such as cfDNA, gDNA, FFPE DNA, etc., NadPrep DNA Library Preparation Module v2 can achieve stable and efficient library yield, achieving high compatibility in different application scenarios.
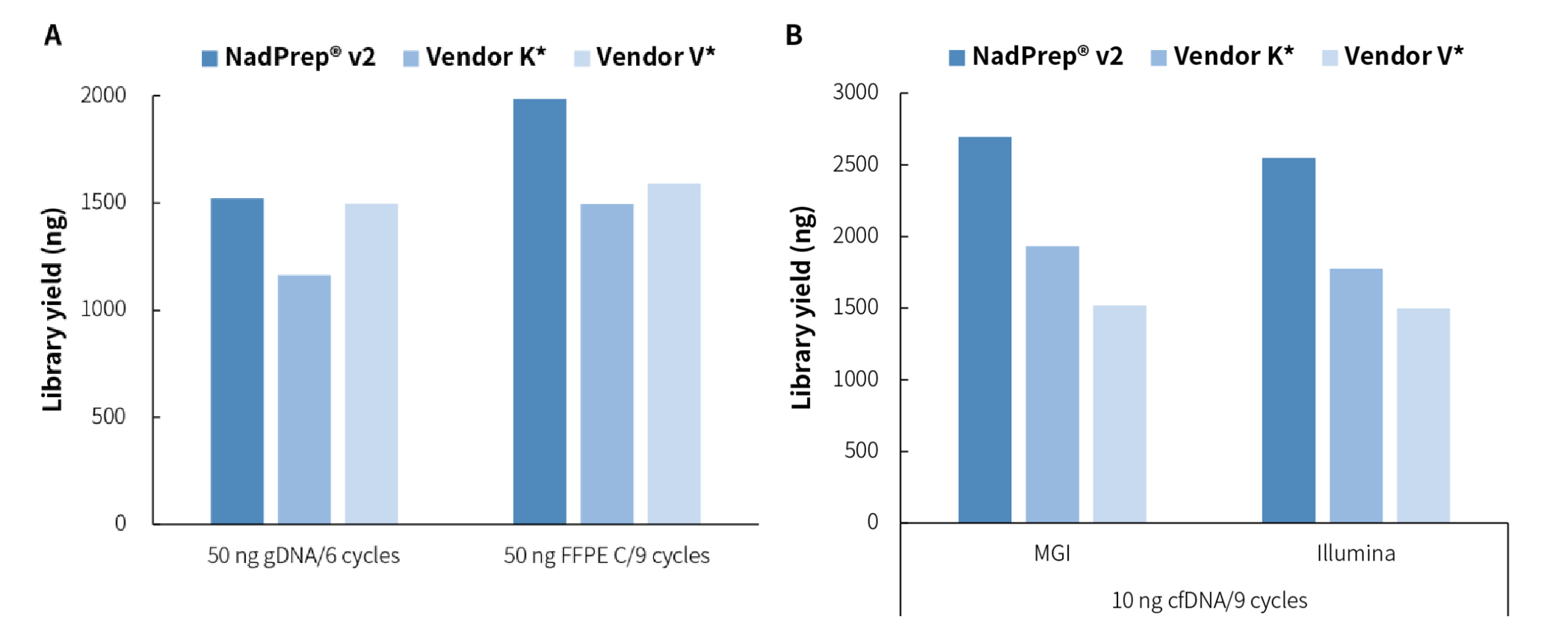 Figure 4. NadPrep DNA Library Preparation Kit v2 has stable and excellent library output for different types of samples. The libraries were prepared by using NadPrep DNA Library Preparation Module v2 coupled with A. NadPrep Universal Stubby Adapter (UDI) Module and B. NadPrep BMI Adapter (MDI) Module (for MGI), and NadPrep UMI Adapter Kit (with 10 nt Index), respectively. The number of amplification cycles followed the corresponding user manuals.
Figure 4. NadPrep DNA Library Preparation Kit v2 has stable and excellent library output for different types of samples. The libraries were prepared by using NadPrep DNA Library Preparation Module v2 coupled with A. NadPrep Universal Stubby Adapter (UDI) Module and B. NadPrep BMI Adapter (MDI) Module (for MGI), and NadPrep UMI Adapter Kit (with 10 nt Index), respectively. The number of amplification cycles followed the corresponding user manuals.
3.1.3 Compatible with different types of adapters
When NadPrep DNA Library Preparation Module v2 is used in combination with different types of adapters for library preparation, stable library yield can be achieved. Compared to the v1 version, NadPrep DNA Library Preparation Module v2 has undergone targeted optimization, resulting in a significant increase in library yield (Figure 5.).
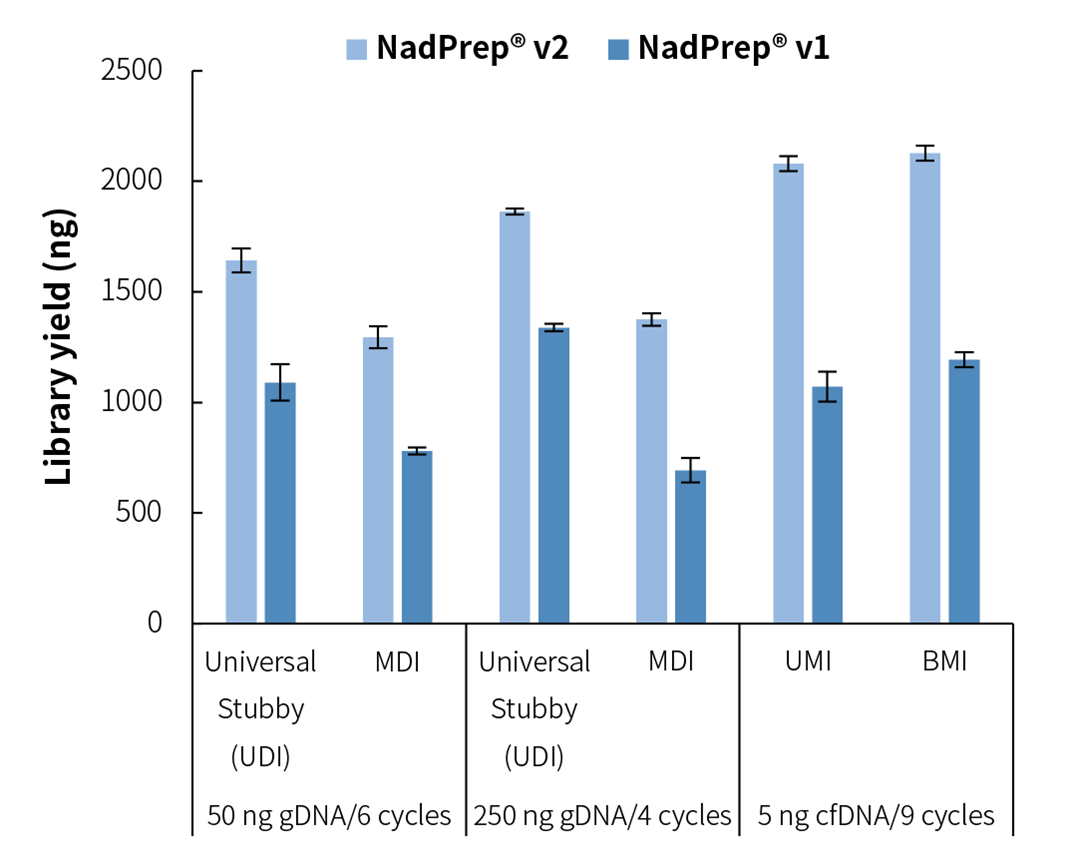
Figure 5. Library yield of NadPrep DNA Library Preparation Kit v2 with different types of adapters. The libraries were prepared from different input samples by using NadPrep DNA Library Preparation Kit/Module (for Illumina®/MGI) (NadPrep v1) and NadPrep DNA Library Preparation Module v2 (NadPrep v2), coupled with NadPrep Universal Stubby Adapter (UDI) Module [Universal Stubby (UDI)], NadPrep Universal Adapter (MDI) Module (for MGI) [MDI], NadPrep UMI Adapter Kit (with 10 nt Index) [UMI], and NadPrep BMI Adapter (MDI) Module (for MGI) [BMI].
3.2 Low GC Bias
We compared the GC bias of NadPrep DNA Library Preparation Module v2 with Vendor K* & V* using microbial community DNA standards with different GC contents (Zymo Research, D6306). The results showed that NadPrep DNA Library Preparation Module v2 exhibited low GC bias.
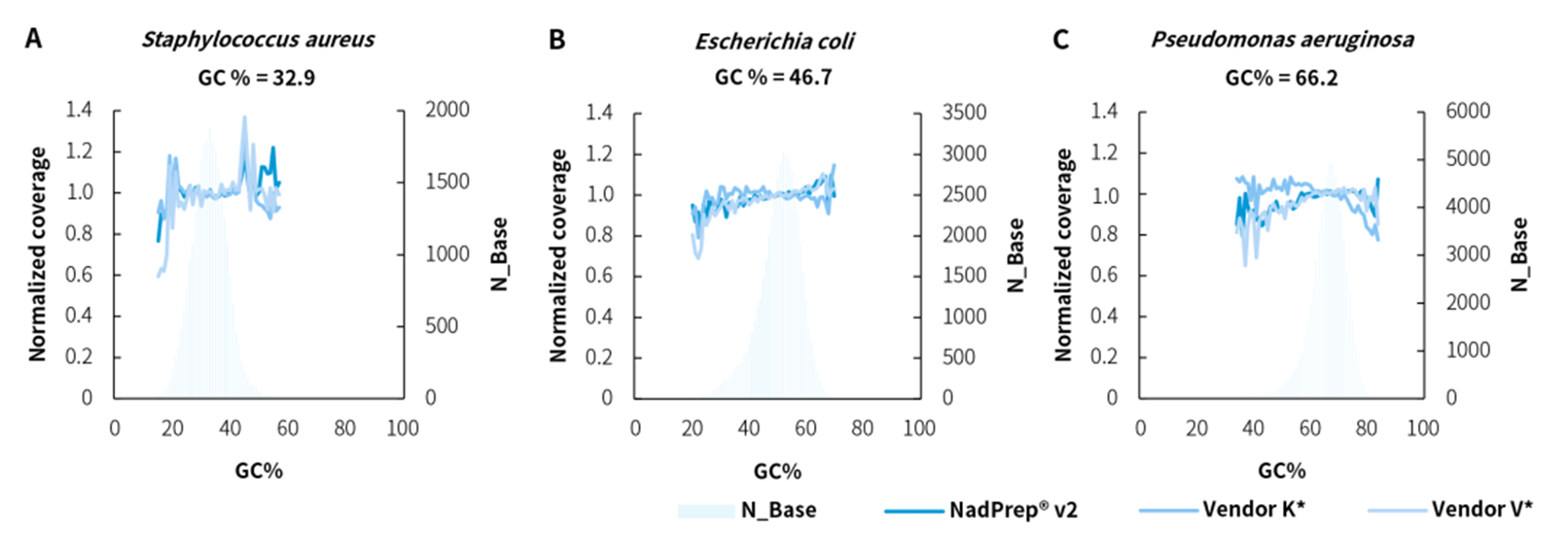
Figure 6. Low GC bias in metagenomic library preparation using NadPrep DNA Library Preparation Kit v2 samples. GC bias of whole-genome libraries with various GC content such as A. Staphylococcus aureus, B. Escherichia coli, and C. Pseudomonas aeruginosa.
3.3 Higher Library Conversion Rate
Library conversion rate is an important indicator for evaluating library quality, as it directly affects the complexity and quality of the library, thereby having a significant impact on the subsequent sequencing analysis results. High conversion rate is particularly important during the identification of ultra-low-frequency mutations in cfDNA samples, as higher conversion rates mean greater library complexity and coverage. Compared with the v1 version, NadPrep DNA Library Preparation Module v2 has been carefully optimized to significantly improve the library conversion rate; In addition, NadPrep DNA Library Preparation Module v2 has a higher library conversion rate compared to Vendor K* & V*, further highlighting its advantages in library preparation efficiency and quality.
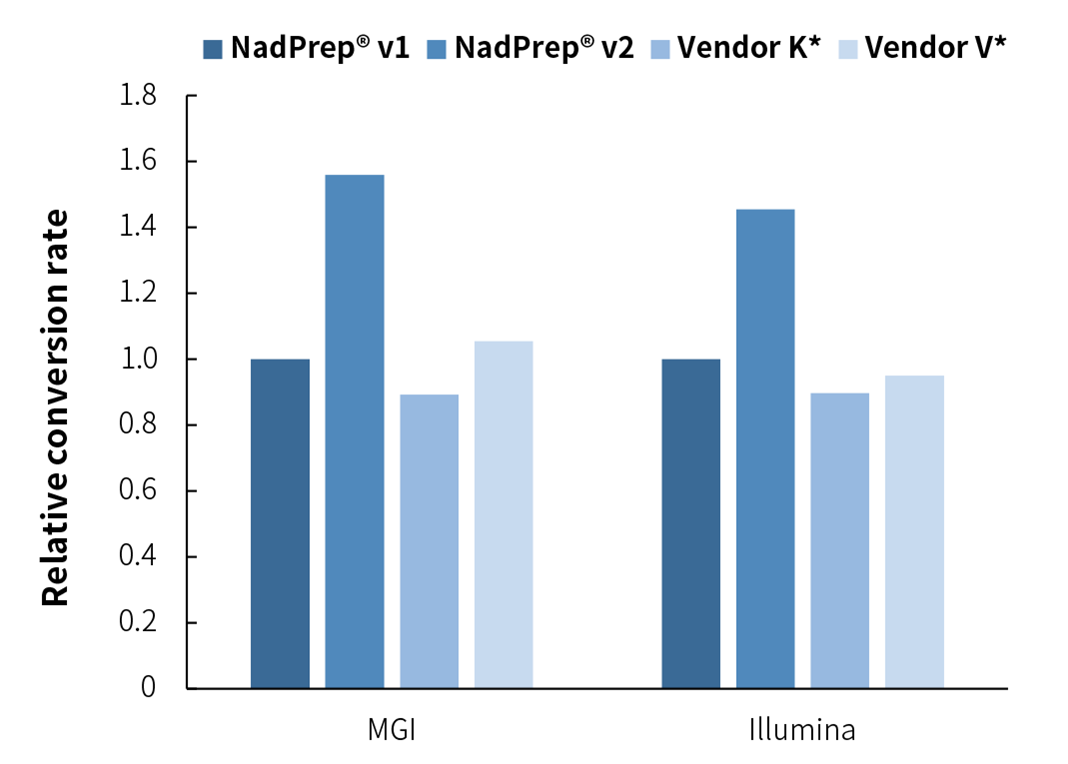
Figure 7. NadPrep DNA Library Preparation Module v2 has higher library conversion rate. 10 ng plasma cfDNA were prepared by using NadPrep DNA Library Preparation Module v2 coupled with NadPrep BMI Adapter (MDI) Module (for MGI) and NadPrep UMI Adapter Kit (with 10 nt Index). The capture was performed by using a 2 Kb Panel with μCaler Hybrid System. Sequencing platform: DNBSEQ-T7, PE150; Illumina Novaseq 6000, PE150. For each sample, 1.0 Gb of data was randomly selected for data analysis, and filtering was performed based on Duplex Consensus Sequences (DCS211).
Note: The relative library conversion rate refers to the ratio of library conversion rates achieved by NadPrep v2 and competitor library preparation kits to that of NadPrep DNA Library Preparation Kit/Module (for Illumina®/MGI) (NadPrep v1), with the library conversion rate of NadPrep v1 normalized to 1).
3.4 Higher Library Complexity
Likewise, to assess library complexity, pre-libraries containing adapters with paired-end unique molecular identifiers were prepared using 10 ng cfDNA from plasma samples and we compared the performance between NadPrep DNA Library Preparation Module v2 and Vendor K* & V* based on the depth of double-stranded consensus sequence coverage. The results showed that the library complexity of NadPrep DNA Library Preparation Module v2 was higher than that of Vendor K* & V*.
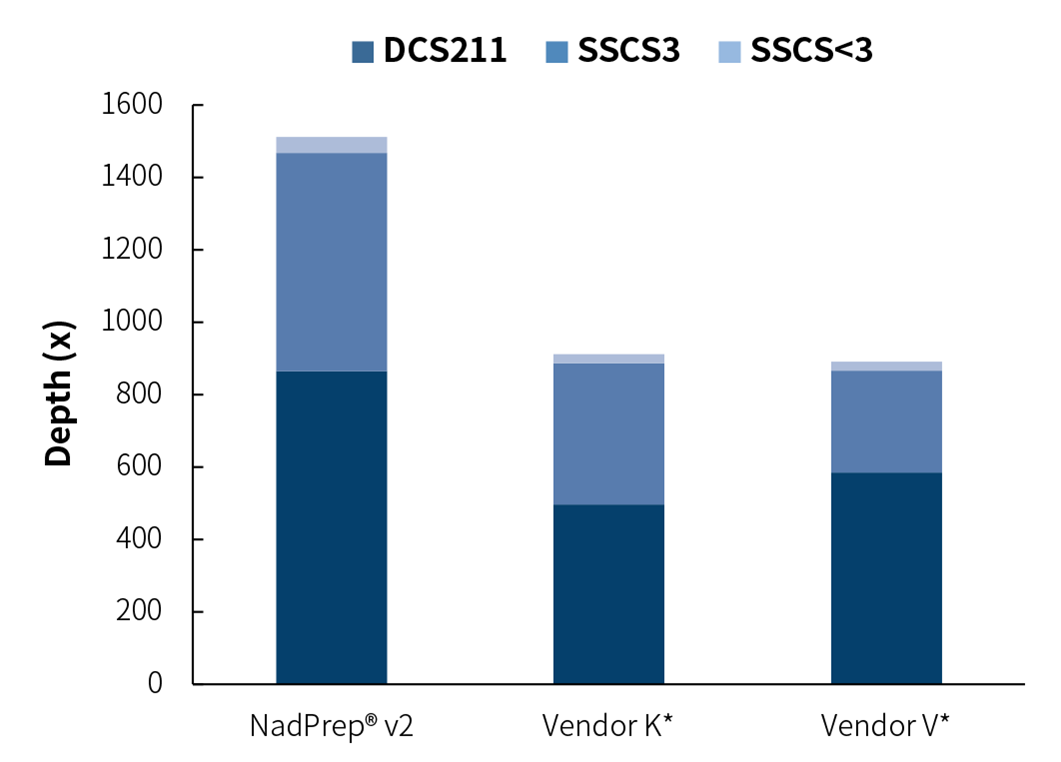
Figure 8. NadPrep DNA Library Preparation Module v2 has higher complexity. Library was prepared using NadPrep DNA Library Preparation Module v2 coupled with NadPrep UMI Adapter Kit (with 10 nt Index). The capture was performed using a 2 Kb Panel with μCaler hybrid system. Sequencing platform: Illumina Novaseq 6000, PE150. For each sample, 1.0 Gb of data was randomly selected for data analysis, and filtering was performed based on Duplex Consensus Sequences (DCS211).
3.5 Compatible with Liquid-phase Hybridization Capture
3.6 Analysis of Mutation in Standard
Table 1. Consistency analysis of standard variation frequency
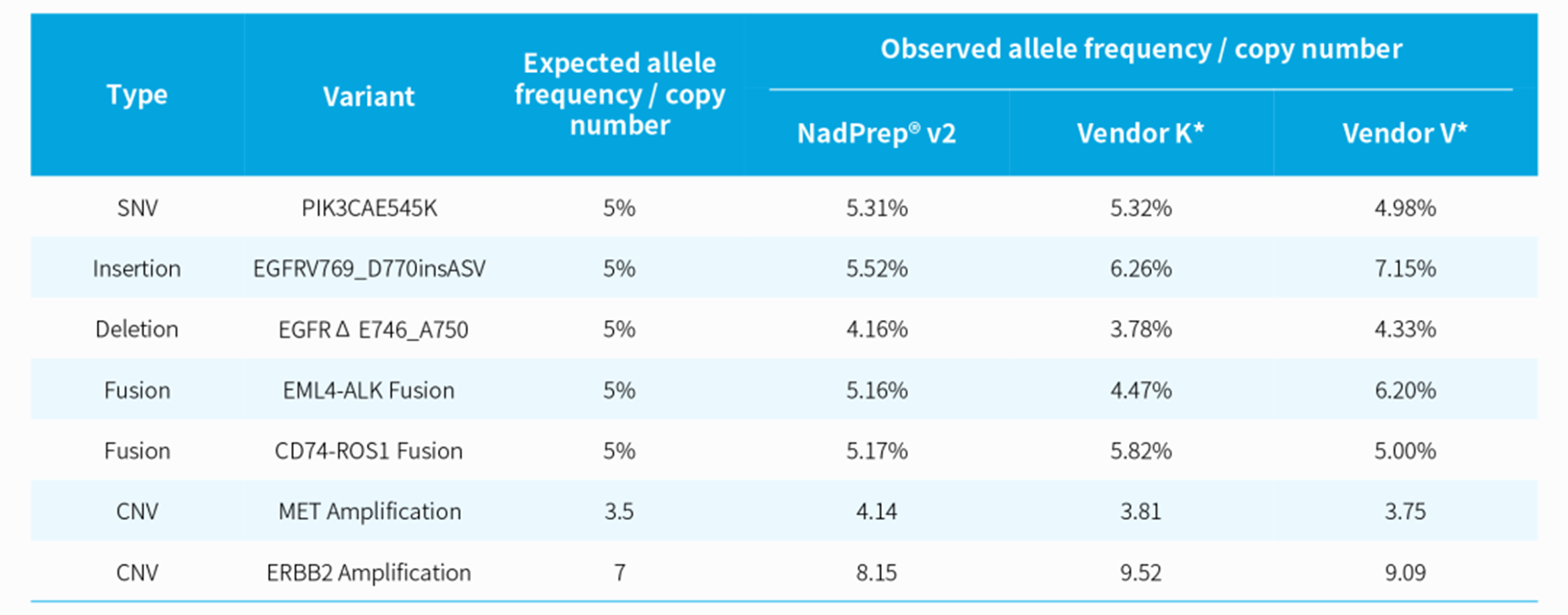
The libraries were prepared by using NadPrep DNA Library Preparation Kit v2 coupled with NadPrep Universal Stubby Adapter (UDI) Module, with 6 cycles of amplification. The capture was performed using NadPrep Hybrid Capture Reagents with the LungCancer Panel v1.0 (0.23 Mb), with a hybridization time of 16 hr. Sequencing platform: Illumina Novaseq 6000, PE 150.
3.7 Freeze-thaw Stability and Accelerated Stability
After 15 and 20 repeated freeze-thaw cycles of all the reagent components, s slightly decrease in library yield was observed, which still meets quality control requirements compared to the starting point (0 cycle). (Figure 10. A & B). Additionally, no significant difference in library yield were observed after all reagent components being left at room temperature for 3, 7, and 14 days compared to the starting point (0 day) (Figure 10. C & D).
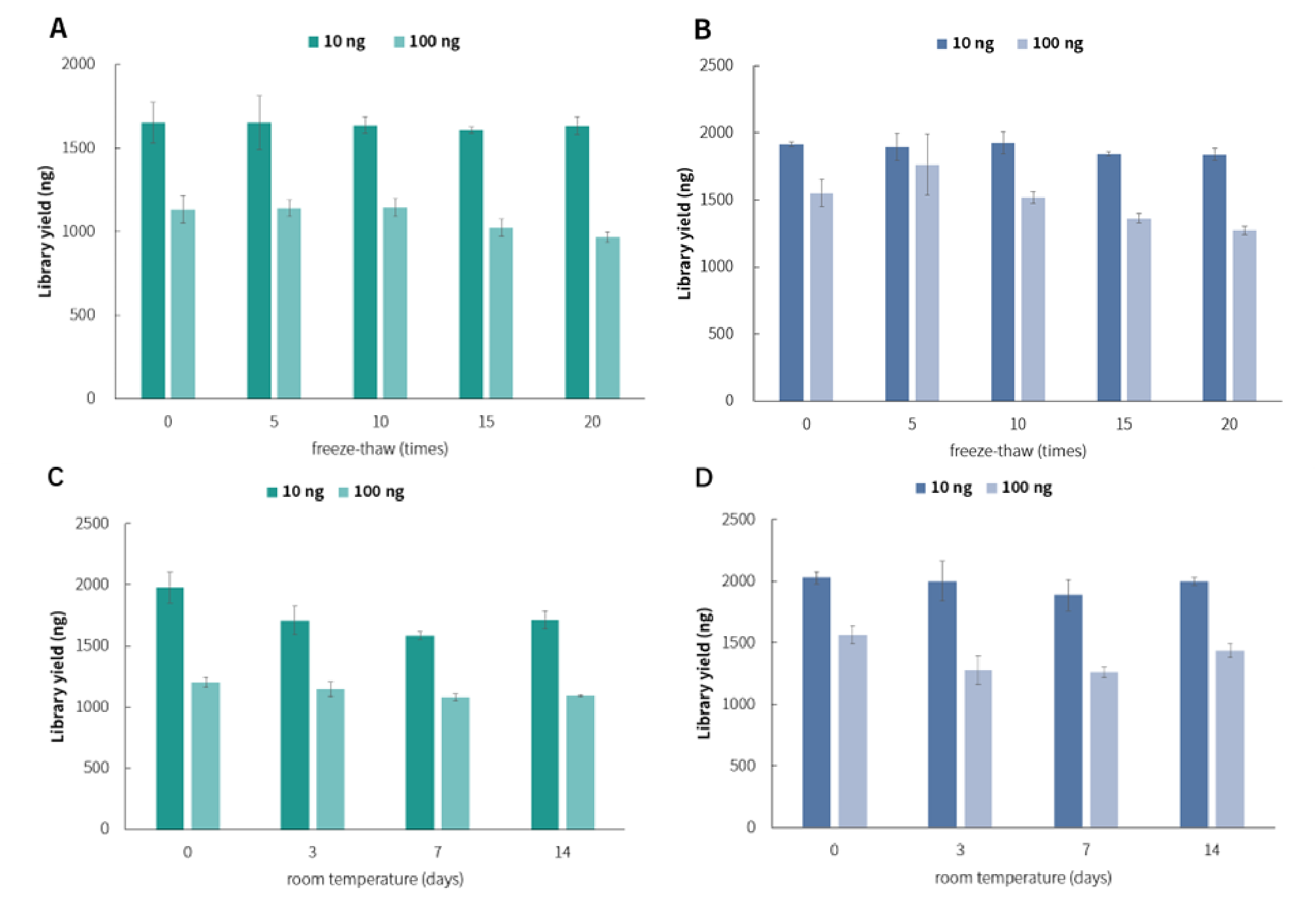
Figure 10. The impact of repeated freeze-thaw cycles and temperature storage of reagents on library yield. Libraries were prepared by using NadPrep DNA Library Preparation Module v2 coupled with NadPrep Universal Adapter (MDI) Module (for MGI) and NadPrep Universal Stubby Adapter (UDI) Module. Sequencing platform: DNBSEQ-T7, PE 150; Illumina® Novaseq 6000, PE 150. A & C. Library yield (MGI); B & D. Library yield (Illumina).
Note: gDNA samples are human genomic DNA standard (Promega, G1471), with an input amount of 10 ng amplified for 9 cycles, and an input amount of 100 ng amplified for 5 cycles.
04 Summary
In summary, NadPrep DNA Library Preparation Module v2 exhibits excellent library preparation performance for various sample types, such as cfDNA, gDNA, FFPE DNA,etc. By improving the library conversion rate, NadPrep DNA Library Preparation Module v2 achieves higher library yield and complexity while maintaining advantages such as low GC bias. Additionally, this kit can be flexibly paired with various types of adapters launched by Nanodigmbio, aiming to provide users with a more stable, effective, accurate, and efficient NGS library preparation solution.
DNA library preparation is the first step in any Next Generation Sequencing (NGS) workflow and is the process whereby DNA or RNA are processed prior to sequencing. It begins with fragmentation, either mechanical or enzymatic to shear the genomic DNA from the target samples into the appropriate insert size. Subsequently, the ends of the DNA are then repaired and primed for the ligation of specific sequencing adapters which are added to the 3’ and 5’ ends. This process results in the formation of an effective library that meets the requirements of the sequencing platform.
The entire process of a routine NGS experiment includes nucleic acid extraction, library preparation, target capture, sequencing, and data analysis. Among them, library preparation, as a core step in the wet-lab NGS workflow, is crucial for the successful conduct of the NGS process. Therefore, the quality of the library prepared using different library preparation kits directly affects the accuracy and reliability of the final sequencing results. With the continuous advancement of NGS technology, NGS has been widely applied in various fields such as basic research, genetic disease detection, non-invasive prenatal testing (NIPT), tumor companion diagnostics (CDx), pre-implantation genetic screening (PGS), and identification of infectious pathogens. Users have placed higher demands for the stability, effectiveness, accuracy, and efficiency of sequencing libraries. From this perspective, it is particularly important to improve the convenience of the NGS library preparation process and the conversion rate (the proportion of initial DNA successfully converted into a sequencable library).
Therefore, in order to meet market demands, Nanodigmbio has launched an upgraded library preparation kit: NadPrep DNA Library Preparation Module v2, which can complete whole-genome library preparation within 3 hours. Compared to the v1 version, the v2 version has improved the library yield and better tolerance for low-quality samples, as well as increased the library conversion rate. Additionally, NadPrep DNA Library Preparation Module v2 is compatible with various types of adapters for different sequencing platforms already launched by Nanodigmbio, allowing for flexible and free combinations.
02 Upgraded Version Library Preparation Kit
2.1 Introduction
NadPrep DNA Library Preparation Module v2 is designed for the preparation of high-quality libraries from double-stranded DNA (dsDNA) on Illumina® & MGI platforms. This A-T ligation-based kit offers a stable and efficient library preparation solution for applications including whole genome sequencing of 1‒500 ng DNA and hybridization capture based targeted sequencing. Compared to the v1 version, the upgraded kit has improved the library conversion efficiency and better tolerance for low-quality samples. Consequently, it can enhance library complexity while maintaining high fidelity.

Figure 1. Entire experimental process of library preparation with NadPrep DNA Library Preparation Module v2.
2.2 Features

Figure 2. Features of NadPrep DNA Library Preparation Module v2.
03 Performance
3.1 Stable and Efficient Library Yield
3.1.1 Compatible with a wide range of input amounts
Using the human genomic DNA standard (Promega, G1521) as a template with different initial input amounts, library preparation was performed using NadPrep DNA Library Preparation Module v2. The results indicate that the library yield at each initial input amount meets the expected output, demonstrating the suitability of this kit for sample initial input amounts ranging from 1 ng to 500 ng.
 Figure 3. Library yield of NadPrep DNA Library Preparation Kit v2 for gDNA samples with different input amounts. The libraries were prepared by using NadPrep DNA Library Preparation Module v2 coupled with NadPrep Universal Adapter (MDI) Module (for MGI) and NadPrep Universal Stubby Adapter (UDI) Module. A. Library yield (MGI); B. Library yield (Illumina).
Figure 3. Library yield of NadPrep DNA Library Preparation Kit v2 for gDNA samples with different input amounts. The libraries were prepared by using NadPrep DNA Library Preparation Module v2 coupled with NadPrep Universal Adapter (MDI) Module (for MGI) and NadPrep Universal Stubby Adapter (UDI) Module. A. Library yield (MGI); B. Library yield (Illumina).Note: gDNA samples are human genomic DNA standard (Promega, G1471).
3.1.2 Compatible with various types of samples
For different types of samples, such as cfDNA, gDNA, FFPE DNA, etc., NadPrep DNA Library Preparation Module v2 can achieve stable and efficient library yield, achieving high compatibility in different application scenarios.
 Figure 4. NadPrep DNA Library Preparation Kit v2 has stable and excellent library output for different types of samples. The libraries were prepared by using NadPrep DNA Library Preparation Module v2 coupled with A. NadPrep Universal Stubby Adapter (UDI) Module and B. NadPrep BMI Adapter (MDI) Module (for MGI), and NadPrep UMI Adapter Kit (with 10 nt Index), respectively. The number of amplification cycles followed the corresponding user manuals.
Figure 4. NadPrep DNA Library Preparation Kit v2 has stable and excellent library output for different types of samples. The libraries were prepared by using NadPrep DNA Library Preparation Module v2 coupled with A. NadPrep Universal Stubby Adapter (UDI) Module and B. NadPrep BMI Adapter (MDI) Module (for MGI), and NadPrep UMI Adapter Kit (with 10 nt Index), respectively. The number of amplification cycles followed the corresponding user manuals.3.1.3 Compatible with different types of adapters
When NadPrep DNA Library Preparation Module v2 is used in combination with different types of adapters for library preparation, stable library yield can be achieved. Compared to the v1 version, NadPrep DNA Library Preparation Module v2 has undergone targeted optimization, resulting in a significant increase in library yield (Figure 5.).

Figure 5. Library yield of NadPrep DNA Library Preparation Kit v2 with different types of adapters. The libraries were prepared from different input samples by using NadPrep DNA Library Preparation Kit/Module (for Illumina®/MGI) (NadPrep v1) and NadPrep DNA Library Preparation Module v2 (NadPrep v2), coupled with NadPrep Universal Stubby Adapter (UDI) Module [Universal Stubby (UDI)], NadPrep Universal Adapter (MDI) Module (for MGI) [MDI], NadPrep UMI Adapter Kit (with 10 nt Index) [UMI], and NadPrep BMI Adapter (MDI) Module (for MGI) [BMI].
3.2 Low GC Bias
We compared the GC bias of NadPrep DNA Library Preparation Module v2 with Vendor K* & V* using microbial community DNA standards with different GC contents (Zymo Research, D6306). The results showed that NadPrep DNA Library Preparation Module v2 exhibited low GC bias.

Figure 6. Low GC bias in metagenomic library preparation using NadPrep DNA Library Preparation Kit v2 samples. GC bias of whole-genome libraries with various GC content such as A. Staphylococcus aureus, B. Escherichia coli, and C. Pseudomonas aeruginosa.
3.3 Higher Library Conversion Rate
Library conversion rate is an important indicator for evaluating library quality, as it directly affects the complexity and quality of the library, thereby having a significant impact on the subsequent sequencing analysis results. High conversion rate is particularly important during the identification of ultra-low-frequency mutations in cfDNA samples, as higher conversion rates mean greater library complexity and coverage. Compared with the v1 version, NadPrep DNA Library Preparation Module v2 has been carefully optimized to significantly improve the library conversion rate; In addition, NadPrep DNA Library Preparation Module v2 has a higher library conversion rate compared to Vendor K* & V*, further highlighting its advantages in library preparation efficiency and quality.

Figure 7. NadPrep DNA Library Preparation Module v2 has higher library conversion rate. 10 ng plasma cfDNA were prepared by using NadPrep DNA Library Preparation Module v2 coupled with NadPrep BMI Adapter (MDI) Module (for MGI) and NadPrep UMI Adapter Kit (with 10 nt Index). The capture was performed by using a 2 Kb Panel with μCaler Hybrid System. Sequencing platform: DNBSEQ-T7, PE150; Illumina Novaseq 6000, PE150. For each sample, 1.0 Gb of data was randomly selected for data analysis, and filtering was performed based on Duplex Consensus Sequences (DCS211).
Note: The relative library conversion rate refers to the ratio of library conversion rates achieved by NadPrep v2 and competitor library preparation kits to that of NadPrep DNA Library Preparation Kit/Module (for Illumina®/MGI) (NadPrep v1), with the library conversion rate of NadPrep v1 normalized to 1).
3.4 Higher Library Complexity
Likewise, to assess library complexity, pre-libraries containing adapters with paired-end unique molecular identifiers were prepared using 10 ng cfDNA from plasma samples and we compared the performance between NadPrep DNA Library Preparation Module v2 and Vendor K* & V* based on the depth of double-stranded consensus sequence coverage. The results showed that the library complexity of NadPrep DNA Library Preparation Module v2 was higher than that of Vendor K* & V*.

Figure 8. NadPrep DNA Library Preparation Module v2 has higher complexity. Library was prepared using NadPrep DNA Library Preparation Module v2 coupled with NadPrep UMI Adapter Kit (with 10 nt Index). The capture was performed using a 2 Kb Panel with μCaler hybrid system. Sequencing platform: Illumina Novaseq 6000, PE150. For each sample, 1.0 Gb of data was randomly selected for data analysis, and filtering was performed based on Duplex Consensus Sequences (DCS211).
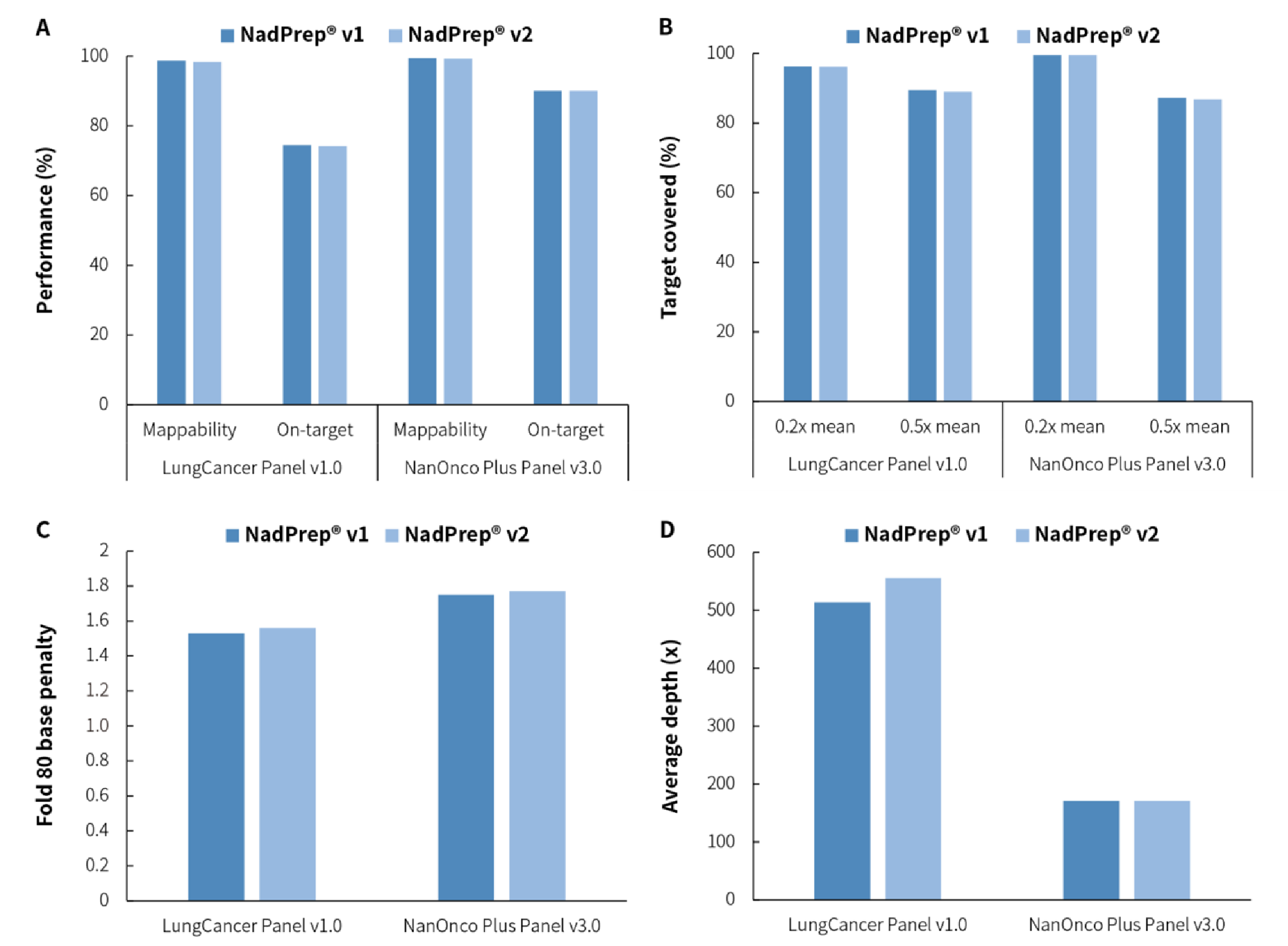
3.5 Compatible with Liquid-phase Hybridization Capture
When NadPrep DNA Library Preparation Module v2 was applied to two panels of different sizes for targeted capture, it showed excellent performance in terms of capture data, such as mappability, on-target rate, coverage uniformity, Fold 80, etc., continuing the excellent targeted capture performance of v1.
3.6 Analysis of Mutation in Standard
Table 1. Consistency analysis of standard variation frequency

The libraries were prepared by using NadPrep DNA Library Preparation Kit v2 coupled with NadPrep Universal Stubby Adapter (UDI) Module, with 6 cycles of amplification. The capture was performed using NadPrep Hybrid Capture Reagents with the LungCancer Panel v1.0 (0.23 Mb), with a hybridization time of 16 hr. Sequencing platform: Illumina Novaseq 6000, PE 150.
3.7 Freeze-thaw Stability and Accelerated Stability
After 15 and 20 repeated freeze-thaw cycles of all the reagent components, s slightly decrease in library yield was observed, which still meets quality control requirements compared to the starting point (0 cycle). (Figure 10. A & B). Additionally, no significant difference in library yield were observed after all reagent components being left at room temperature for 3, 7, and 14 days compared to the starting point (0 day) (Figure 10. C & D).

Figure 10. The impact of repeated freeze-thaw cycles and temperature storage of reagents on library yield. Libraries were prepared by using NadPrep DNA Library Preparation Module v2 coupled with NadPrep Universal Adapter (MDI) Module (for MGI) and NadPrep Universal Stubby Adapter (UDI) Module. Sequencing platform: DNBSEQ-T7, PE 150; Illumina® Novaseq 6000, PE 150. A & C. Library yield (MGI); B & D. Library yield (Illumina).
Note: gDNA samples are human genomic DNA standard (Promega, G1471), with an input amount of 10 ng amplified for 9 cycles, and an input amount of 100 ng amplified for 5 cycles.
04 Summary
In summary, NadPrep DNA Library Preparation Module v2 exhibits excellent library preparation performance for various sample types, such as cfDNA, gDNA, FFPE DNA,etc. By improving the library conversion rate, NadPrep DNA Library Preparation Module v2 achieves higher library yield and complexity while maintaining advantages such as low GC bias. Additionally, this kit can be flexibly paired with various types of adapters launched by Nanodigmbio, aiming to provide users with a more stable, effective, accurate, and efficient NGS library preparation solution.
Solutions
- Methyl Library Preparation Total Solution
- Sequencing single library on different platform--Universal Stubby Adapter (UDI)
- HRD score Analysis
- Unique Dual Index for MGI platforms
- RNA-Cap Sequencing of Human Respiratory Viruses Including SARS-CoV-2
- Total Solution for RNA-Cap Sequencing
- Total Solution for MGI Platforms
- Whole Exome Sequencing
- Low-frequency Mutation Analysis
Events
-
Exhibition Preview | Nanodigmbio invites you to join us at Boston 2025 Annual Meeting of the American Society of Human Genetics (ASHG)

-
Exhibition Preview | Nanodigmbio Invites You to Join Us at WHX & WHX Labs Kuala Lumpur 2025, Malaysia International Trade and Exhibition Centre in Kuala Lumpur

-
Exhibition Preview | Nanodigmbio Invites You to Join Us at Hospitalar 2025, Brazil International Medical Device Exhibition in São Paulo

-
Exhibition Preview | Nanodigmbio invites you to join us at Denver 2024 Annual Meeting of the American Society of Human Genetics (ASHG)

-
Exhibition Preview | Nanodigmbio invites you to join us at Sapporo 2024 Annual Meeting of the Japan Society of Human Genetics (JSHG)

-
Exhibition Preview | Nanodigmbio invites you to join us at Association for Diagnostics & Laboratory Medicine (ADLM)