New Arrival | μCaler EMS Panel v1.0: Precise Detection of Multigene Methylation, Screening for 9 Major High-Incidence Cancers
View: 5351 / Time: 2024-09-02
01 Background
Cancer early screening can effectively increase the proportion of early-stage cancer patients (apparently healthy population), enabling patients to receive interventions and treatment earlier, thereby reducing mortality and improving the five-year survival rate of cancer patients. However, from a global perspective, compared to widely accepted health check-ups, the acceptance and prevalence of cancer early screening and detection are still relatively low. Taking colorectal cancer as an example, the five-year survival rate for stage I (early stage) patients can exceed 90%, while for stage IV (late stage) patients, it drops to 14% [1]. Traditional tumor biomarkers may have slightly lower sensitivity and accuracy in detectable rate, and are often limited to a single type of cancer, which are limiting factors for the promotion of cancer early screening. Compared to potential biomarkers for cancer early screening such as proteins, fragmentomics, small molecule metabolites, miRNA, and ctDNA mutations, DNA methylation occurs in almost all precancerous lesions and early stages of cancer. DNA methylation biomarkers not only exhibit diversity in modification sites but also possess tissue/cancer specificity, along with significant advantages in signal abundance and intensity, making them one of the most widely applicable multi-cancer early screening biomarkers currently available.
In November 2023, Nanodigmbio launched the world's first μCaler Hybrid System for Methylation. This system utilizes the internationally patented exclusive technology (μCaler), providing users with a precise, stable, efficient, fast, and simple methylation sequencing experience. It not only enhances the efficiency and accuracy of methylation detection but also represents a new breakthrough for multi-cancer early screening.
To fully leverage the advantages of the μCaler Methylation Hybrid Capture System in the field of cancer early screening, Nanodigmbio has launched the μCaler EMS Panel v1.0 (Early Methylation for Screening, hereinafter referred to as "EMS"). EMS combined with the latest upgraded μCaler Hybrid Capture Reagents v2 and μCaler NanoBlockers, forms a comprehensively optimized methylation early screening technical solution. This solution can accurately capture and convert methylated libraries, comprehensively covering the methylation status targets of candidate genes for target cancer types in the tested samples, providing strong support for the early cancer screening and detection.
02 Introduction
2.1 μCaler EMS Panel
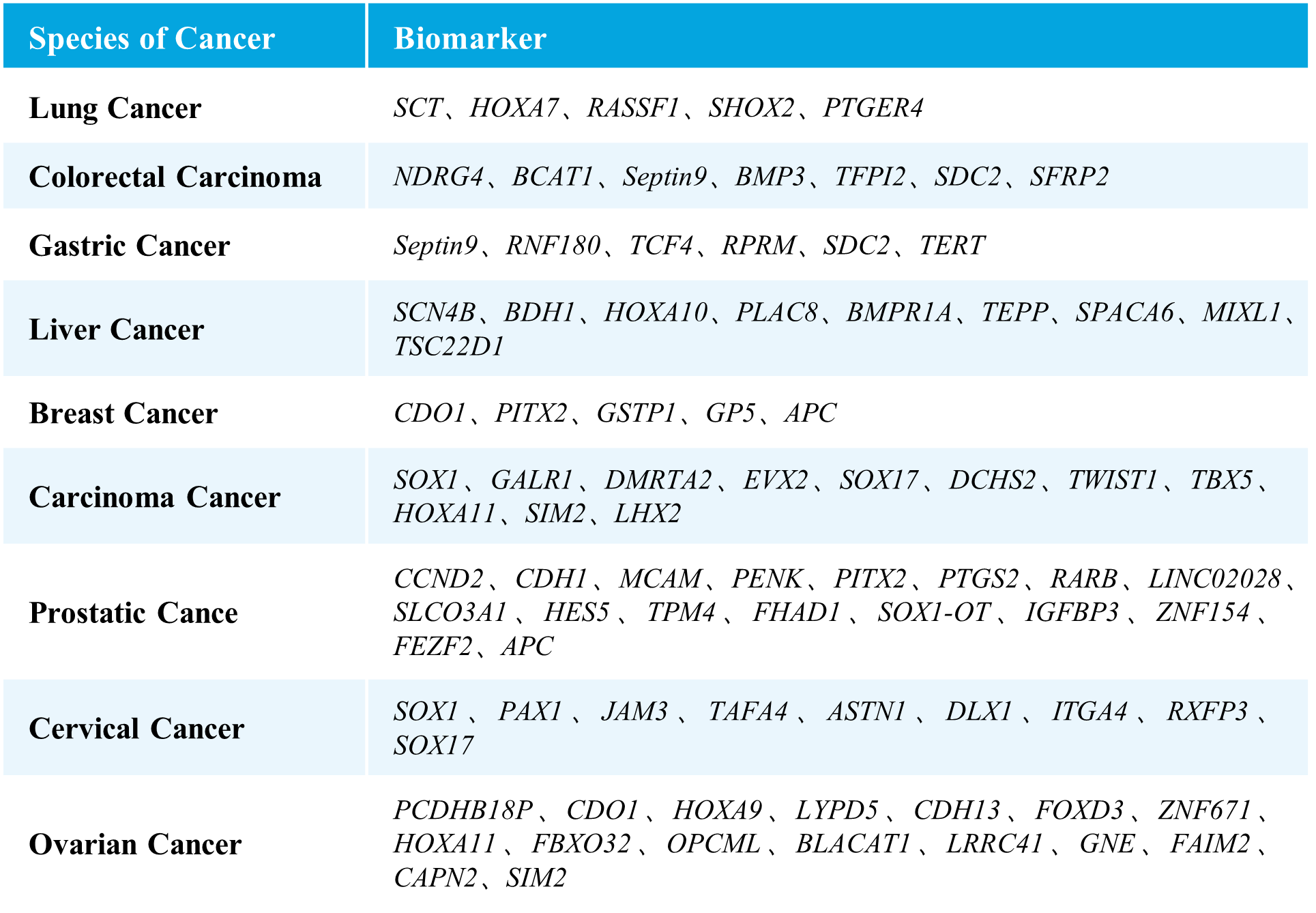
μCaler EMS Panel v1.0 covers methylation gene sites related to nine major high-incidence cancers, including selected sites approved by the NMPA and FDA, as well as those reported in literature and patents. It encompasses nine types of cancers, including lung cancer, colorectal cancer, gastric cancer, liver cancer, breast cancer, carcinoma cancer, prostate cancer, cervical cancer, and ovarian cancer, involving 76 methylated candidate genes associated with carcinogenesis and tumor suppression, with over 2,000 CpG sites. The probe design covers approximately 20 Kb of the human genome, providing comprehensive and accurate support for methylation early screening.
Table 1. List of Cancer Types and Genes Covered in μCaler EMS Panel v1.0.
 2.2 Features
2.2 Features
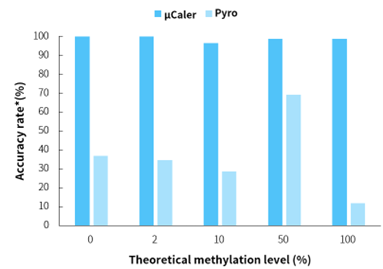
Figure 1. μCaler significantly enhances the accuracy of methylation level quantification.
Note: *Evaluation criteria for simulated samples with different methylation levels are: 0 ± 5%, 2 ± 5%, 10 ± 5%, 50 ± 5%, 100 ± 15%. Accuracy rate is the proportion of sites meeting the evaluation criteria.
03 Performance
3.1 Compatibility with Different Sample Types
3.1.1 gDNA & cfDNA
Using human genomic DNA standards and cfDNA from plasma samples, methylated libraries were performed for hybrid capture with the μCaler EMS Panel v1.0. For gDNA samples, the capture performance is illustrated in Figure 2. A & C, with an average on-target rate exceeding 60%. The mean coverage for CpG sites at 0.2x depth reached over 99%, and even at 0.5x depth, it remained above 95%, demonstrating excellent coverage of all target CpG sites, with a Fold 80 value controlled below 1.5. The capture performance of plasma cfDNA samples is shown in Figure 2. B & C. During the conversion step, whether using bisulfite (BS) or enzyme (EM), the average on-target rate could reach over 55%. The mean coverage for CpG sites at 0.2x depth was consistently higher than 99%, and at 0.5x depth, it remained above 95%,with a Fold 80 value controlled below 1.8.
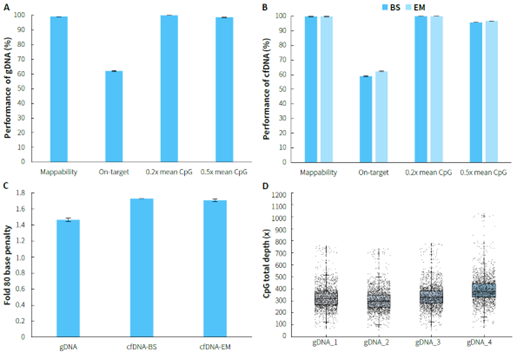
Figure 2. Capture performance of μCaler EMS Panel v1.0 applied to gDNA and cfDNA samples. A. gDNA ; B. cfDNA; C. Fold 80 base penalty; D. CpG total depth of target regions (deduplicated). Samples were used to prepare pre-library using the NadPrep Methyl Library Preparation Module, with gDNA undergoing conversion using NadPrep DNA Methyl Bisulfite Conversion Module (BS), and cfDNA undergoing conversion using both BS and enzymatic (EM) methods. 500 ng/pre-library was inputted, and hybrid capture was performed using μCaler Hybrid Capture Reagents v2 and μCaler EMS Panel v1.0.
Note: gDNA samples refer to Human Genomic DNA standard (Promega, G1471), with an input of 50 ng; cfDNA samples refer to plasma cfDNA, with an input of 20 ng.
3.1.2 FFPE DNA
FFPE samples are widely used in clinical pathology testing and tumor genetic analysis due to their ability to preserve tissues for long periods or prepare specimens for examination. However, the poor quality of nucleic acids in FFPE samples often affects the quality of library preparation, posing a significant challenge. Performing methylated DNA libraries preparation using different grades of FFPE samples, followed by hybrid capture with μCaler EMS Panel v1.0. As shown in Figure 3, EMS can effectively detect different grades of FFPE DNA samples, with mean coverage for CpG sites at 0.2x depth exceeding 98%. Even for D-grade FFPE samples, the mean coverage for CpG sites at 0.5x depth can reach above 80%.
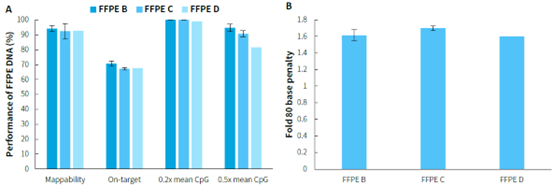
Figure 3. Capture performance of μCaler EMS Panel v1.0 applied to different grades of FFPE samples. A. Capture performance; B. Fold 80 base penalty. Pre-library were prepared using the NadPrep Methyl Library Preparation Module, followed by conversion using the NadPrep DNA Methyl Bisulfite Conversion Module. 500 ng of pre-library was inputted, and hybrid capture was performed using μCaler Hybrid Capture Reagents v2 and μCaler EMS Panel v1.0.
Note: FFPE samples were clinical lung cancer samples, input 50 ng. Sample grading criteria: FFPE B: Main bands around 15 Kb with moderate smearing; FFPE C: Bands primarily distributed between 200-2500 bp, appearing diffuse; FFPE D: Bands mainly distributed between 100-1000 bp, appearing diffuse.
3.2 Clinical Sample
3.2.1 Colorectal Cancer
Clinical paired samples from colorectal cancer were used to assess the detection performance of μCaler EMS Panel v1.0. The results showed that the mean coverage for CpG sites at 0.2x depth was above 99% in both tumor tissues and peritumoral tissue (Figure 4.B). Significant differences in methylation levels were observed between the four pairs of samples: the methylation levels in tumor tissues were significantly higher than those in peritumoral tissue (Figure 5.A & B). Seven candidate biomarkers for colorectal cancer have been approved by the NMPA or FDA, and their methylation levels in tumor tissues were generally higher than those in peritumoral tissues. However, some sites also exhibited high methylation levels in peritumoral tissue (Figure 5.B), which may be correlated with the sampling of peritumoral tissue. In addition, the methylation status of candidate biomarkers for colorectal cancer varied among different individuals, indicating the importance of multi-site methylation detection for early screening of colorectal cancer.
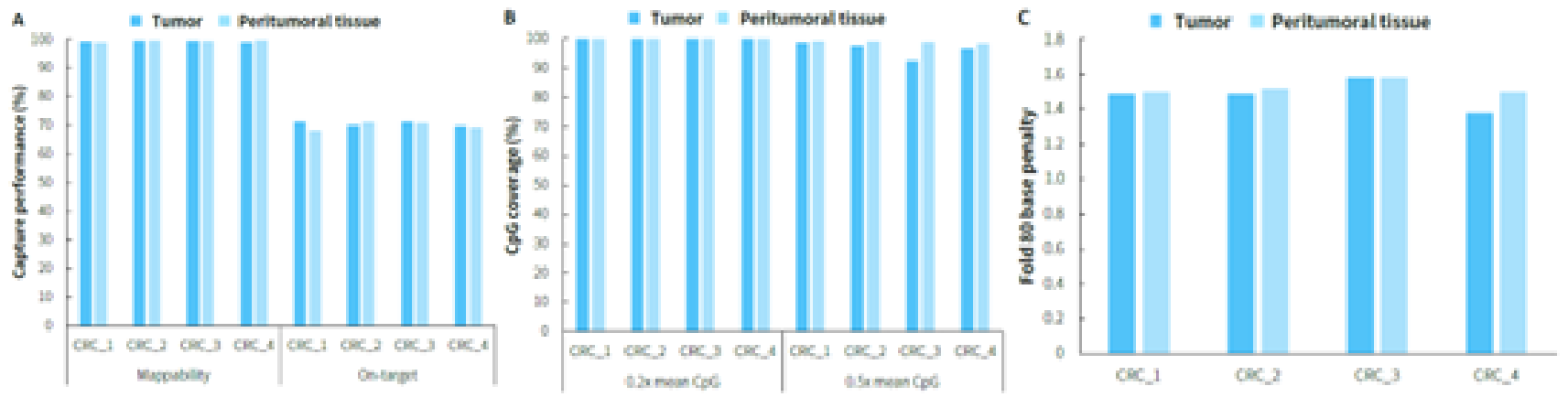 Figure 4. Capture performance of μCaler EMS Panel v1.0 applied to clinical colorectal cancer and peritumoral tissue samples. A. On target & Mappability; B. CpG coverage; C. Fold 80 base penalty.
Figure 4. Capture performance of μCaler EMS Panel v1.0 applied to clinical colorectal cancer and peritumoral tissue samples. A. On target & Mappability; B. CpG coverage; C. Fold 80 base penalty.
Note: CRC: Colorectal Cancer.
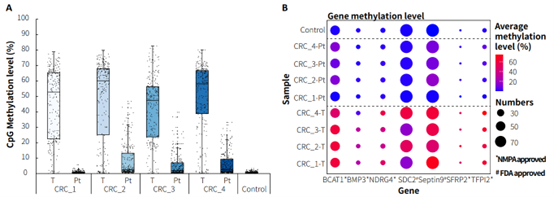
Figure 5. Methylation level detection of μCaler EMS Panel v1.0 applied to clinical colorectal cancer and peritumoral tissue samples. A. CpG methylation level in paired colorectal cancer samples; B. CpG methylation level of colorectal cancer candidate biomarkers in real samples.
Note: CRC: Colorectal Cancer; Pt: Peritumoral tissue.
3.2.2 Ovarian Cancer
Additionally, we validated the detection performance of μCaler EMS Panel v1.0 using clinical paired samples from ovarian cancer. The results showed that the mean coverage for CpG sites at 0.2x depth exceeded 99%, and the mean coverage at 0.5x depth exceeded 95%, demonstrating excellent coverage (Figure 6.B). Among the three paired samples, the methylation levels in tumor tissues were significantly higher than those in the corresponding peritumoral tissues and control samples (Promega, G1471) (Figure 7.A), indicating that EMS can effectively detect differences in methylation levels between tumor and peritumoral tissue.
Most of the candidate biomarkers for ovarian cancer exhibited high methylation levels in tumor tissues and low methylation levels in peritumoral tissue (Figure 7.B). However, there were also some biomarkers that, while showing higher methylation levels in tumor tissues compared to peritumoral tissues, still exhibit relatively high methylation levels in peritumoral tissues and control samples, such as FOXD3, LYPD5, PCDHB18P (Figure 7.B). Additionally, we found similar situations for these three genes in other cancer types. Therefore, further validation and screening based on more clinical data are needed to determine whether these biomarkers are suitable for early screening of ovarian cancer.
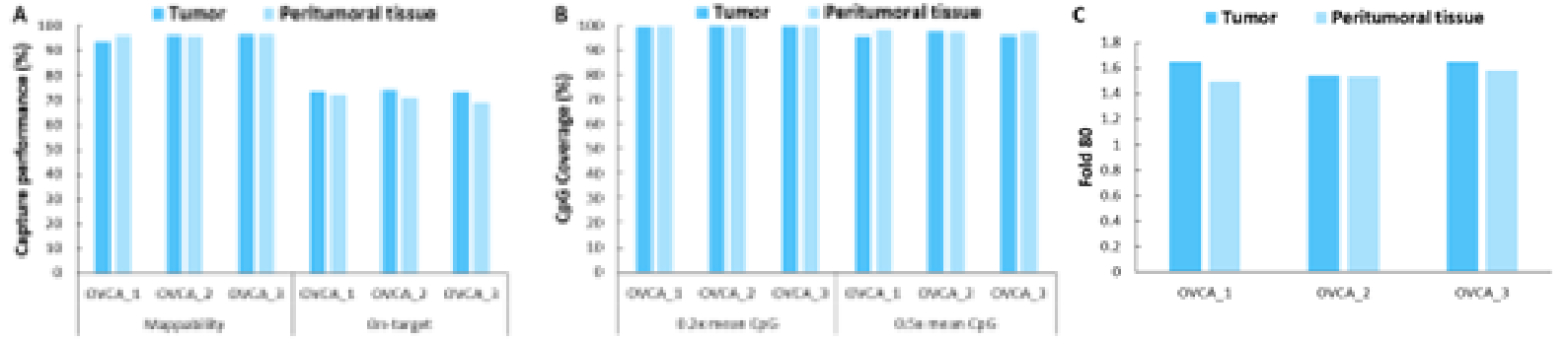 Figure 6. Capture performance of μCaler EMS Panel v1.0 applied to clinical ovarian cancer and peritumoral tissue samples. A. On target & Mappability; B. CpG coverage; C. Fold 80 base penalty.
Figure 6. Capture performance of μCaler EMS Panel v1.0 applied to clinical ovarian cancer and peritumoral tissue samples. A. On target & Mappability; B. CpG coverage; C. Fold 80 base penalty.
Note: OVCA: Ovarian Cancer.
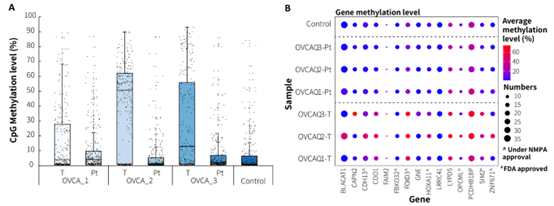
Figure 7. The methylation level detection of μCaler EMS Panel v1.0 applied to clinical ovarian cancer and peritumoral tissue. A. CpG methylation level in paired ovarian cancer samples; B. CpG methylation level of ovarian cancer candidate biomarkers in real samples.
Note: OVCA: Ovarian Cancer; Pt: Peritumoral tissue.
04 Application Prospects
DNA Methylation, as an important epigenetic biomarker, currently demonstrates tremendous application prospects in various aspects related to cancer, including tumor early screening/ diagnosis, molecular subtyping, prognosis stratification, treatment efficacy assessment, and more. Therefore, the establishment and screening of biomarkers have become the main research directions. μCaler EMS Panel v1.0 adopts patented probe design and is equipped with the novel μCaler Methylation Hybrid Capture System, applied in the field of multi-cancer methylation detection. Based on the genes and sites covered by this panel, users can choose to supplement the required CpG sites independently, increasing its flexibility and applicability.
Nanodigmbio focuses on providing comprehensive solutions and technical support for clinical cohort studies, biomarker development, and validation. The comprehensive technical solutions will help accelerate the research process of early screening products and make positive contributions to the field of early cancer diagnosis.
[1] Siegel R L, Wagle N S, Cercek A, et al. Colorectal cancer statistics, 2023[J]. CA: a cancer journal for clinicians, 2023, 73(3): 233-254.
Cancer early screening can effectively increase the proportion of early-stage cancer patients (apparently healthy population), enabling patients to receive interventions and treatment earlier, thereby reducing mortality and improving the five-year survival rate of cancer patients. However, from a global perspective, compared to widely accepted health check-ups, the acceptance and prevalence of cancer early screening and detection are still relatively low. Taking colorectal cancer as an example, the five-year survival rate for stage I (early stage) patients can exceed 90%, while for stage IV (late stage) patients, it drops to 14% [1]. Traditional tumor biomarkers may have slightly lower sensitivity and accuracy in detectable rate, and are often limited to a single type of cancer, which are limiting factors for the promotion of cancer early screening. Compared to potential biomarkers for cancer early screening such as proteins, fragmentomics, small molecule metabolites, miRNA, and ctDNA mutations, DNA methylation occurs in almost all precancerous lesions and early stages of cancer. DNA methylation biomarkers not only exhibit diversity in modification sites but also possess tissue/cancer specificity, along with significant advantages in signal abundance and intensity, making them one of the most widely applicable multi-cancer early screening biomarkers currently available.
In November 2023, Nanodigmbio launched the world's first μCaler Hybrid System for Methylation. This system utilizes the internationally patented exclusive technology (μCaler), providing users with a precise, stable, efficient, fast, and simple methylation sequencing experience. It not only enhances the efficiency and accuracy of methylation detection but also represents a new breakthrough for multi-cancer early screening.
To fully leverage the advantages of the μCaler Methylation Hybrid Capture System in the field of cancer early screening, Nanodigmbio has launched the μCaler EMS Panel v1.0 (Early Methylation for Screening, hereinafter referred to as "EMS"). EMS combined with the latest upgraded μCaler Hybrid Capture Reagents v2 and μCaler NanoBlockers, forms a comprehensively optimized methylation early screening technical solution. This solution can accurately capture and convert methylated libraries, comprehensively covering the methylation status targets of candidate genes for target cancer types in the tested samples, providing strong support for the early cancer screening and detection.
02 Introduction
2.1 μCaler EMS Panel
μCaler EMS Panel v1.0 covers methylation gene sites related to nine major high-incidence cancers, including selected sites approved by the NMPA and FDA, as well as those reported in literature and patents. It encompasses nine types of cancers, including lung cancer, colorectal cancer, gastric cancer, liver cancer, breast cancer, carcinoma cancer, prostate cancer, cervical cancer, and ovarian cancer, involving 76 methylated candidate genes associated with carcinogenesis and tumor suppression, with over 2,000 CpG sites. The probe design covers approximately 20 Kb of the human genome, providing comprehensive and accurate support for methylation early screening.
Table 1. List of Cancer Types and Genes Covered in μCaler EMS Panel v1.0.
 2.2 Features
2.2 Features- Multi-Cancer Detection: Covers CpG sites of multiple genes for 9 major cancer types in a single detection
- Sample Compatibility: Suitable for gDNA, cfDNA, and various level of FFPE samples.
- Accurate Quantification: Achieves precise quantification of methylation levels in tested samples.
- Efficient Capture: Combines with μCaler Total Solution for Methylation to ensure uniform data quality, higher capture efficiency and stability
- High Reliability: Utilizes exclusive patented probe design to comprehensively detect methylation status, providing reliable results.
- Stable Delivery: In-house nucleic acid synthesis manufacturing ensures on-hand delivery and guarantees the stability of the delivery chain.
- Flexible Customization: Customizes panels containing different genes and scales of CpG sites for testing specific single/ multiple cancer types.

Figure 1. μCaler significantly enhances the accuracy of methylation level quantification.
Note: *Evaluation criteria for simulated samples with different methylation levels are: 0 ± 5%, 2 ± 5%, 10 ± 5%, 50 ± 5%, 100 ± 15%. Accuracy rate is the proportion of sites meeting the evaluation criteria.
03 Performance
3.1 Compatibility with Different Sample Types
3.1.1 gDNA & cfDNA
Using human genomic DNA standards and cfDNA from plasma samples, methylated libraries were performed for hybrid capture with the μCaler EMS Panel v1.0. For gDNA samples, the capture performance is illustrated in Figure 2. A & C, with an average on-target rate exceeding 60%. The mean coverage for CpG sites at 0.2x depth reached over 99%, and even at 0.5x depth, it remained above 95%, demonstrating excellent coverage of all target CpG sites, with a Fold 80 value controlled below 1.5. The capture performance of plasma cfDNA samples is shown in Figure 2. B & C. During the conversion step, whether using bisulfite (BS) or enzyme (EM), the average on-target rate could reach over 55%. The mean coverage for CpG sites at 0.2x depth was consistently higher than 99%, and at 0.5x depth, it remained above 95%,with a Fold 80 value controlled below 1.8.

Figure 2. Capture performance of μCaler EMS Panel v1.0 applied to gDNA and cfDNA samples. A. gDNA ; B. cfDNA; C. Fold 80 base penalty; D. CpG total depth of target regions (deduplicated). Samples were used to prepare pre-library using the NadPrep Methyl Library Preparation Module, with gDNA undergoing conversion using NadPrep DNA Methyl Bisulfite Conversion Module (BS), and cfDNA undergoing conversion using both BS and enzymatic (EM) methods. 500 ng/pre-library was inputted, and hybrid capture was performed using μCaler Hybrid Capture Reagents v2 and μCaler EMS Panel v1.0.
Note: gDNA samples refer to Human Genomic DNA standard (Promega, G1471), with an input of 50 ng; cfDNA samples refer to plasma cfDNA, with an input of 20 ng.
3.1.2 FFPE DNA
FFPE samples are widely used in clinical pathology testing and tumor genetic analysis due to their ability to preserve tissues for long periods or prepare specimens for examination. However, the poor quality of nucleic acids in FFPE samples often affects the quality of library preparation, posing a significant challenge. Performing methylated DNA libraries preparation using different grades of FFPE samples, followed by hybrid capture with μCaler EMS Panel v1.0. As shown in Figure 3, EMS can effectively detect different grades of FFPE DNA samples, with mean coverage for CpG sites at 0.2x depth exceeding 98%. Even for D-grade FFPE samples, the mean coverage for CpG sites at 0.5x depth can reach above 80%.

Figure 3. Capture performance of μCaler EMS Panel v1.0 applied to different grades of FFPE samples. A. Capture performance; B. Fold 80 base penalty. Pre-library were prepared using the NadPrep Methyl Library Preparation Module, followed by conversion using the NadPrep DNA Methyl Bisulfite Conversion Module. 500 ng of pre-library was inputted, and hybrid capture was performed using μCaler Hybrid Capture Reagents v2 and μCaler EMS Panel v1.0.
Note: FFPE samples were clinical lung cancer samples, input 50 ng. Sample grading criteria: FFPE B: Main bands around 15 Kb with moderate smearing; FFPE C: Bands primarily distributed between 200-2500 bp, appearing diffuse; FFPE D: Bands mainly distributed between 100-1000 bp, appearing diffuse.
3.2 Clinical Sample
3.2.1 Colorectal Cancer
Clinical paired samples from colorectal cancer were used to assess the detection performance of μCaler EMS Panel v1.0. The results showed that the mean coverage for CpG sites at 0.2x depth was above 99% in both tumor tissues and peritumoral tissue (Figure 4.B). Significant differences in methylation levels were observed between the four pairs of samples: the methylation levels in tumor tissues were significantly higher than those in peritumoral tissue (Figure 5.A & B). Seven candidate biomarkers for colorectal cancer have been approved by the NMPA or FDA, and their methylation levels in tumor tissues were generally higher than those in peritumoral tissues. However, some sites also exhibited high methylation levels in peritumoral tissue (Figure 5.B), which may be correlated with the sampling of peritumoral tissue. In addition, the methylation status of candidate biomarkers for colorectal cancer varied among different individuals, indicating the importance of multi-site methylation detection for early screening of colorectal cancer.
 Figure 4. Capture performance of μCaler EMS Panel v1.0 applied to clinical colorectal cancer and peritumoral tissue samples. A. On target & Mappability; B. CpG coverage; C. Fold 80 base penalty.
Figure 4. Capture performance of μCaler EMS Panel v1.0 applied to clinical colorectal cancer and peritumoral tissue samples. A. On target & Mappability; B. CpG coverage; C. Fold 80 base penalty.Note: CRC: Colorectal Cancer.

Figure 5. Methylation level detection of μCaler EMS Panel v1.0 applied to clinical colorectal cancer and peritumoral tissue samples. A. CpG methylation level in paired colorectal cancer samples; B. CpG methylation level of colorectal cancer candidate biomarkers in real samples.
Note: CRC: Colorectal Cancer; Pt: Peritumoral tissue.
3.2.2 Ovarian Cancer
Additionally, we validated the detection performance of μCaler EMS Panel v1.0 using clinical paired samples from ovarian cancer. The results showed that the mean coverage for CpG sites at 0.2x depth exceeded 99%, and the mean coverage at 0.5x depth exceeded 95%, demonstrating excellent coverage (Figure 6.B). Among the three paired samples, the methylation levels in tumor tissues were significantly higher than those in the corresponding peritumoral tissues and control samples (Promega, G1471) (Figure 7.A), indicating that EMS can effectively detect differences in methylation levels between tumor and peritumoral tissue.
Most of the candidate biomarkers for ovarian cancer exhibited high methylation levels in tumor tissues and low methylation levels in peritumoral tissue (Figure 7.B). However, there were also some biomarkers that, while showing higher methylation levels in tumor tissues compared to peritumoral tissues, still exhibit relatively high methylation levels in peritumoral tissues and control samples, such as FOXD3, LYPD5, PCDHB18P (Figure 7.B). Additionally, we found similar situations for these three genes in other cancer types. Therefore, further validation and screening based on more clinical data are needed to determine whether these biomarkers are suitable for early screening of ovarian cancer.
 Figure 6. Capture performance of μCaler EMS Panel v1.0 applied to clinical ovarian cancer and peritumoral tissue samples. A. On target & Mappability; B. CpG coverage; C. Fold 80 base penalty.
Figure 6. Capture performance of μCaler EMS Panel v1.0 applied to clinical ovarian cancer and peritumoral tissue samples. A. On target & Mappability; B. CpG coverage; C. Fold 80 base penalty.Note: OVCA: Ovarian Cancer.

Figure 7. The methylation level detection of μCaler EMS Panel v1.0 applied to clinical ovarian cancer and peritumoral tissue. A. CpG methylation level in paired ovarian cancer samples; B. CpG methylation level of ovarian cancer candidate biomarkers in real samples.
Note: OVCA: Ovarian Cancer; Pt: Peritumoral tissue.
04 Application Prospects
DNA Methylation, as an important epigenetic biomarker, currently demonstrates tremendous application prospects in various aspects related to cancer, including tumor early screening/ diagnosis, molecular subtyping, prognosis stratification, treatment efficacy assessment, and more. Therefore, the establishment and screening of biomarkers have become the main research directions. μCaler EMS Panel v1.0 adopts patented probe design and is equipped with the novel μCaler Methylation Hybrid Capture System, applied in the field of multi-cancer methylation detection. Based on the genes and sites covered by this panel, users can choose to supplement the required CpG sites independently, increasing its flexibility and applicability.
Nanodigmbio focuses on providing comprehensive solutions and technical support for clinical cohort studies, biomarker development, and validation. The comprehensive technical solutions will help accelerate the research process of early screening products and make positive contributions to the field of early cancer diagnosis.
[1] Siegel R L, Wagle N S, Cercek A, et al. Colorectal cancer statistics, 2023[J]. CA: a cancer journal for clinicians, 2023, 73(3): 233-254.
Solutions
- Methyl Library Preparation Total Solution
- Sequencing single library on different platform--Universal Stubby Adapter (UDI)
- HRD score Analysis
- Unique Dual Index for MGI platforms
- RNA-Cap Sequencing of Human Respiratory Viruses Including SARS-CoV-2
- Total Solution for RNA-Cap Sequencing
- Total Solution for MGI Platforms
- Whole Exome Sequencing
- Low-frequency Mutation Analysis
Events
-
Exhibition Preview | Nanodigmbio invites you to join us at Boston 2025 Annual Meeting of the American Society of Human Genetics (ASHG)

-
Exhibition Preview | Nanodigmbio Invites You to Join Us at WHX & WHX Labs Kuala Lumpur 2025, Malaysia International Trade and Exhibition Centre in Kuala Lumpur

-
Exhibition Preview | Nanodigmbio Invites You to Join Us at Hospitalar 2025, Brazil International Medical Device Exhibition in São Paulo

-
Exhibition Preview | Nanodigmbio invites you to join us at Denver 2024 Annual Meeting of the American Society of Human Genetics (ASHG)

-
Exhibition Preview | Nanodigmbio invites you to join us at Sapporo 2024 Annual Meeting of the Japan Society of Human Genetics (JSHG)

-
Exhibition Preview | Nanodigmbio invites you to join us at Association for Diagnostics & Laboratory Medicine (ADLM)