Unfazed by BS Damage: Dual Boost in Library Yield and Sequencing Depth!
View: 3801 / Time: 2025-01-09
01 Background
DNA methylation, as a key epigenetic modification, holds significant value in fields such as cancer research, genetics, epigenetics, stem cell studies etc[1]. As a pivotal step in the DNA methylation sequencing workflow, the quality of methylation library preparation directly impacts the accuracy and sensitivity of detection. With the growing demands of research, the application of methylation sequencing libraries faces higher requirements in stability, precision, and efficiency, driving continuous advancements in related technologies and products to better meet the rigorous standards of scientific research and clinical applications.
02 NadPrep dsDNA Methylation Library Preparation Solution
The newly upgraded NadPrep Methyl Library Preparation Module v2 is suitable for preparing methylation sequencing (Methyl-Seq) libraries from 1-500 ng DNA samples, supporting whole-genome bisulfite sequencing (WGBS) as well as hybrid capture-based targeted methylation sequencing with effective enrichment of targeted libraries. Meanwhile, it is compatible with various types of DNA samples, including gDNA, cfDNA, and FFPE DNA, as well as a wide range of commercially available conversion solutions.
Compared to the v1 version, the upgraded module shortens experimental time, supports lower sample input amounts, and significantly improves library yield and data utilization efficiency, offering users a more efficient, flexible, and reliable methylation sequencing solution.
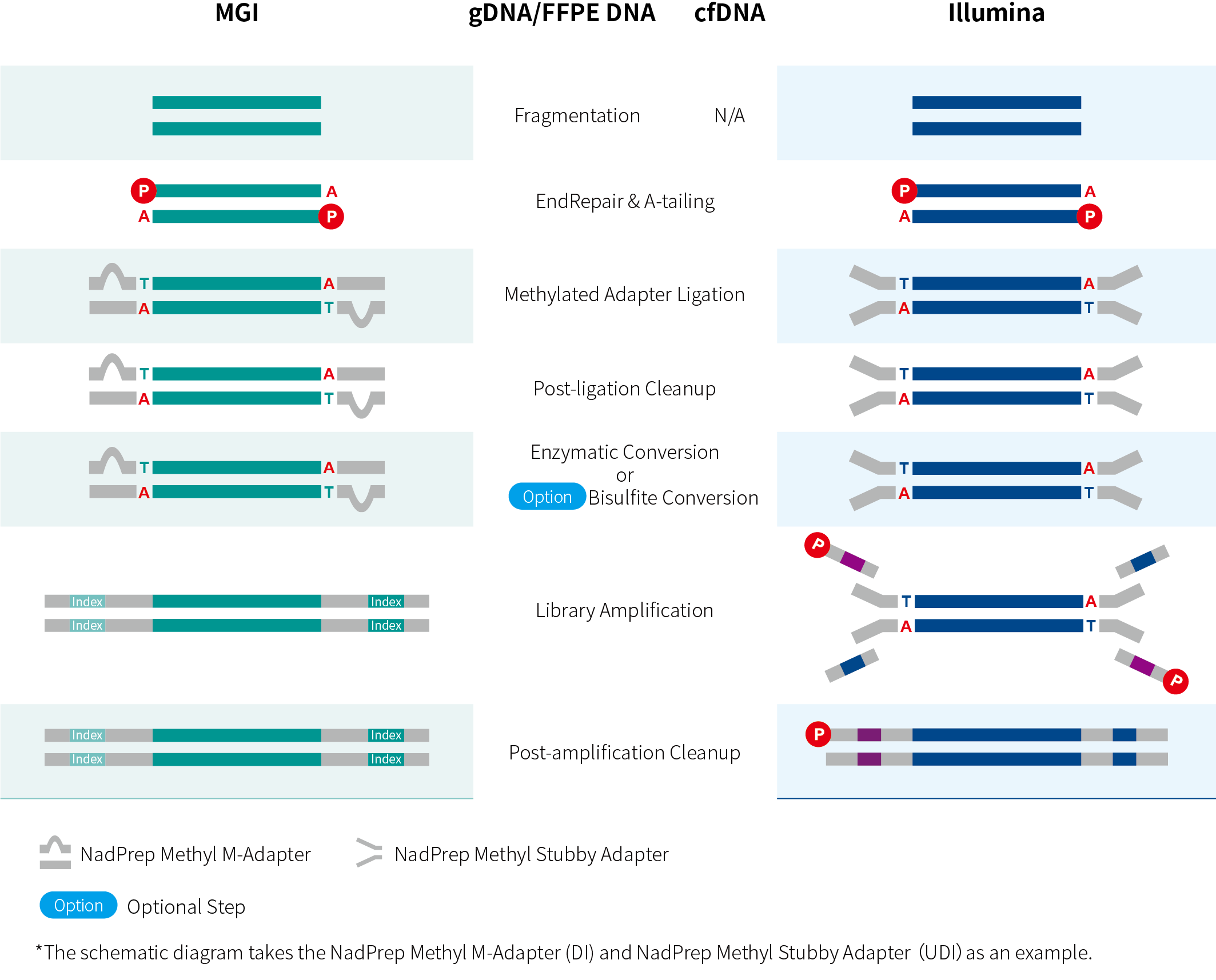
Figure 1. Workflow of NadPrep dsDNA Methylation Library Preparation Solution for MGI and Illumina platforms.
03 Feature
Lower Sample Input: Reduced from 10 ng to as low as 1 ng.
Faster Experimental Workflow: Shortened from 3 hr to 2.5 hr*
Higher Library Yield: Significant improvement in library yield across various applications compared to v1.
Improved Data Utilization: Achieves higher effective sequencing depth than v1, ranking among the best in its class.
Flexible Conversion Options: Supports both bisulfite conversion and enzymatic conversion methods.
Dual Platform Compatibility: Can be applied to both Illumina and MGI sequencing platforms after ligation with different adapters.
*Excluding conversion time.
04 Performance
4.1 Higher Library Yield
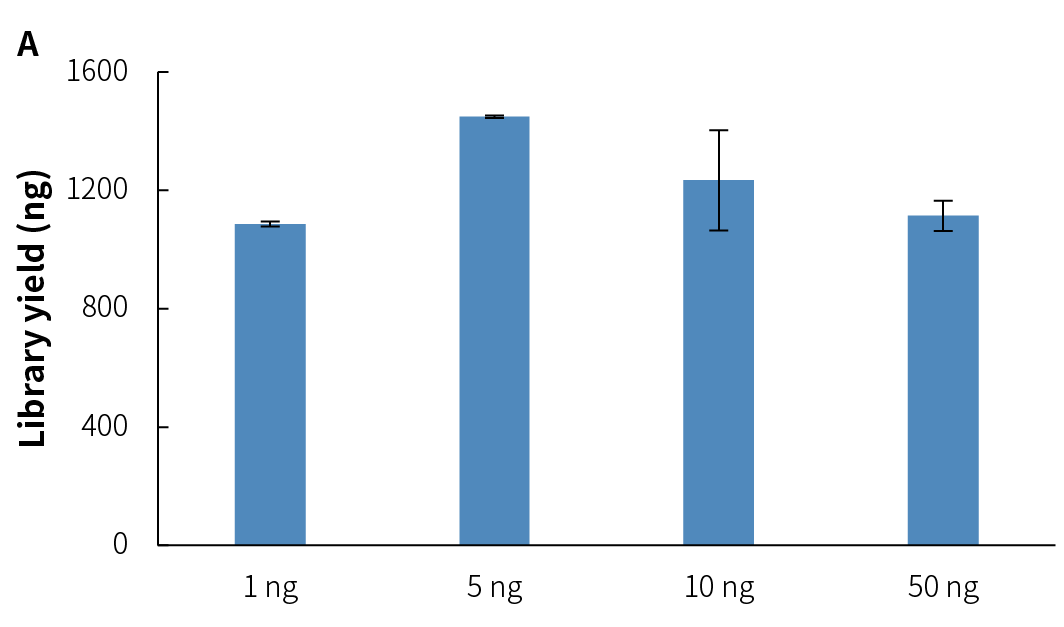
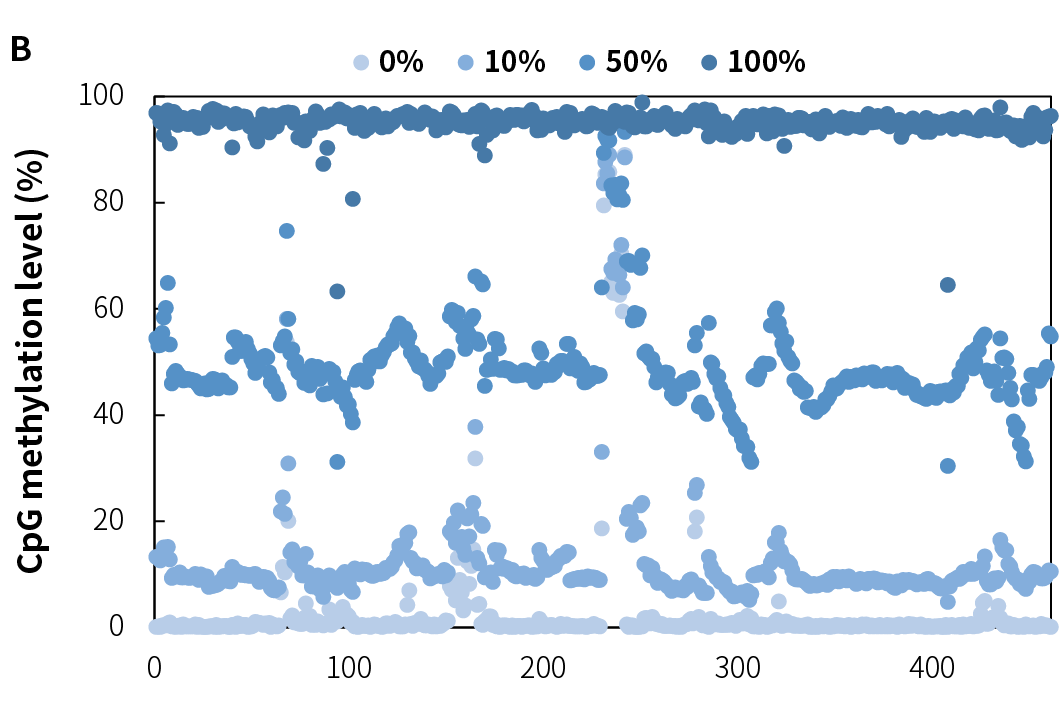
NadPrep Methyl Library Preparation Module v2 (hereinafter referred to as "NadPrep v2") demonstrates consistent library yield across various common sample types, including gDNA, FFPE DNA, and cfDNA (Figure 2.A), as well as simulated samples with different methylation levels (1%, 10%, 50%, and 100%) (Figure 2.B). Compared to v1, NadPrep v2 delivers significantly enhanced library yield across samples with different input amounts (Figure 2.C). This enhanced performance not only improves library yield but also supports compatibility with diverse sample types, enabling more efficient applications in diverse research scenarios.
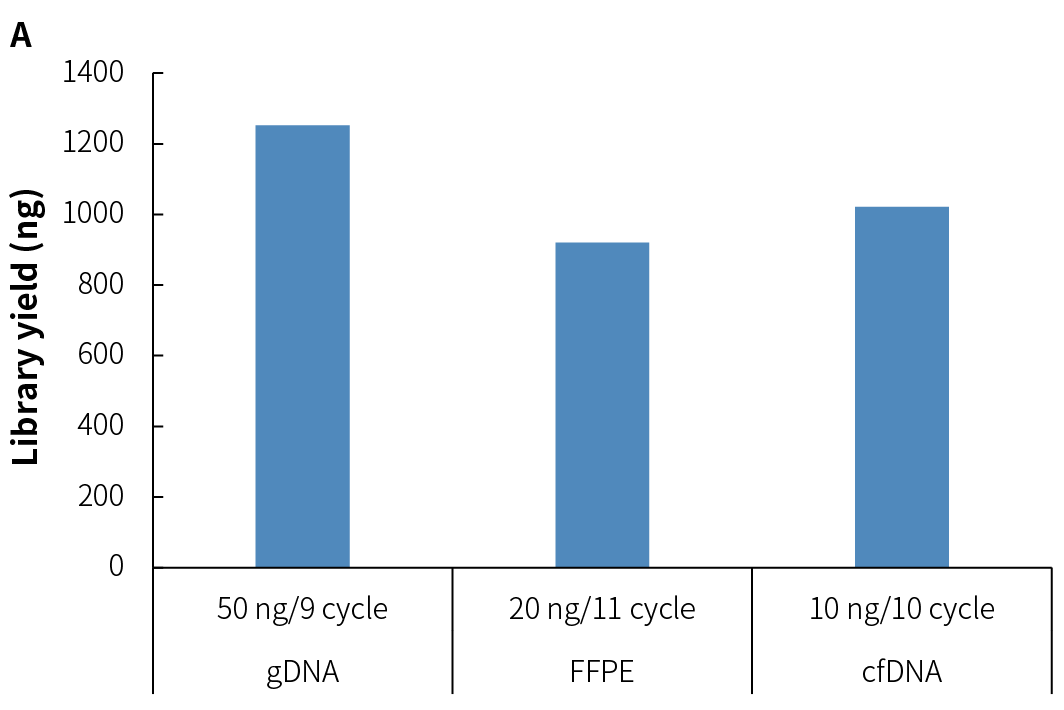
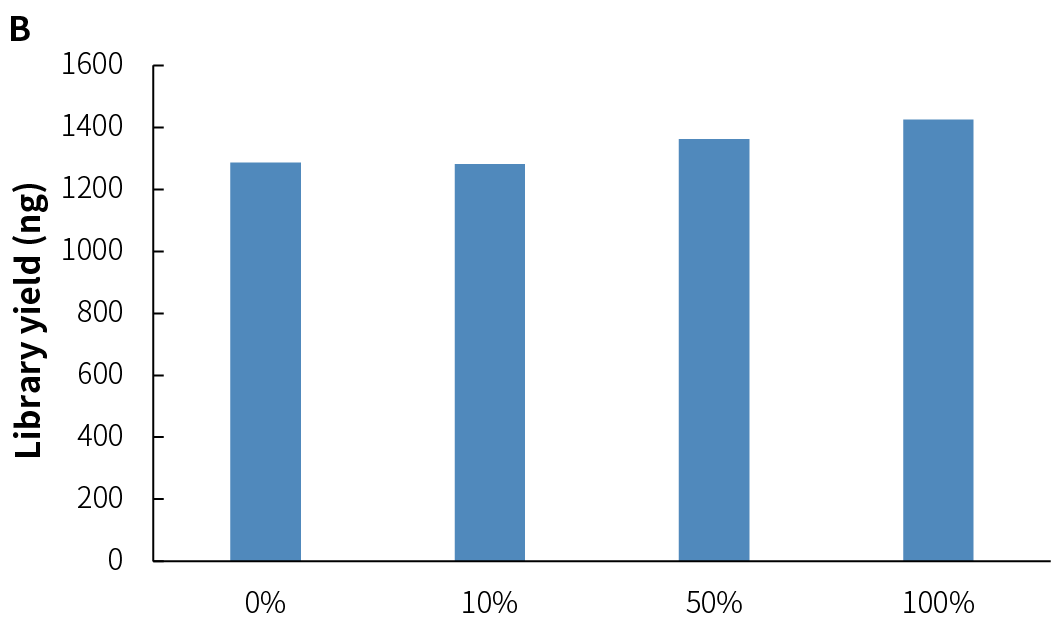
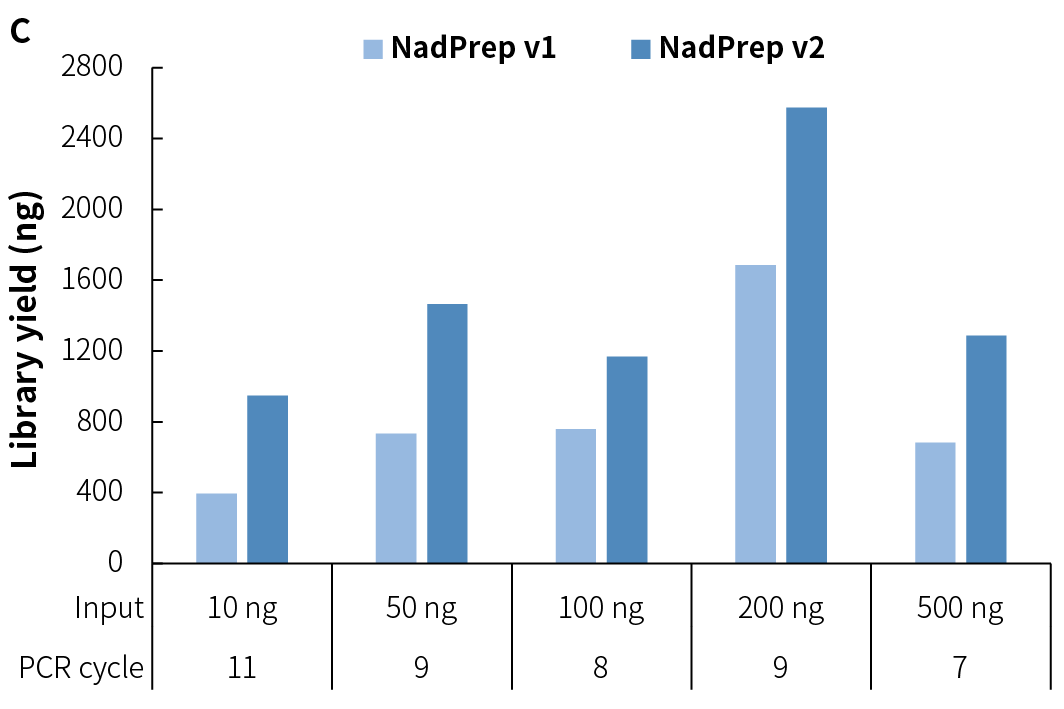
Figure 2. NadPrep Methyl Library Preparation Module v2 ensures stable library yield for different sample types. Libraries were prepared using NadPrep Methyl Library Preparation Module v2 coupled with NadPrep Methyl Stubby Adapter (UDI) Module (with 10 nt Index) and treated by BS conversion. A. gDNA, FFPE DNA, and cfDNA; B. Simulated samples with varying methylation levels; C. gDNA (comparison of v1 vs. v2).
Note: The samples were simulated DNA by using human 100% Methylated DNA standard (Zymo, D5014-2) positive control and 0% Methylated DNA standard (Zymo, D5014-1) negative control to mimic different methylation levels (0%, 10%, and 50%), with an initial input amount of 50 ng.
4.2 Improved Data Utilization
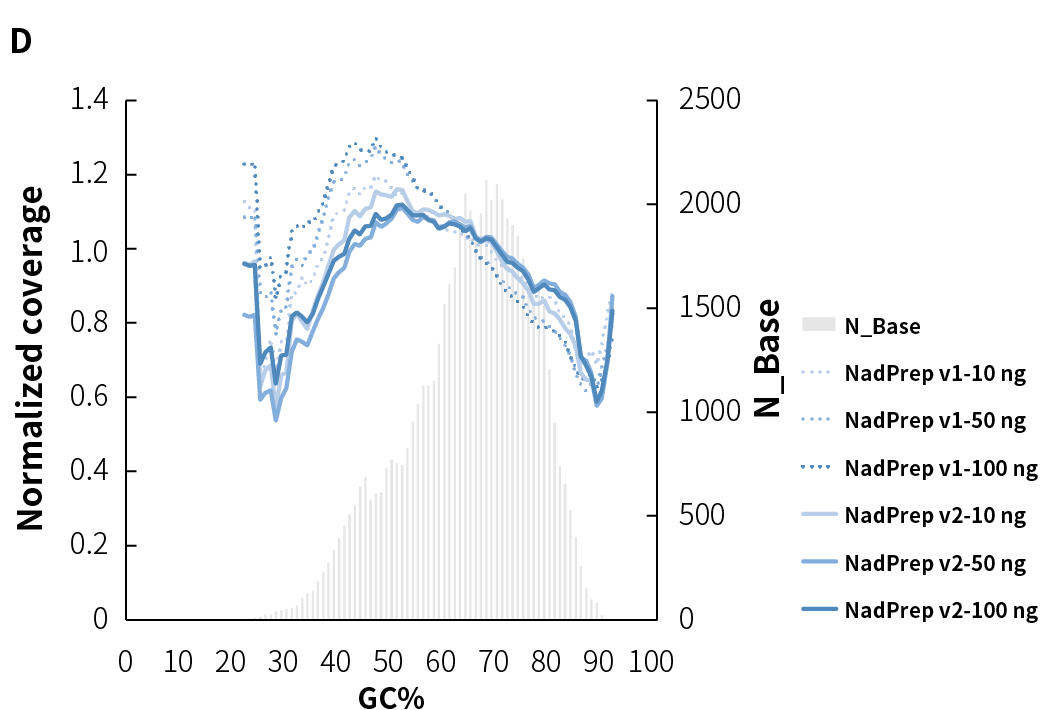
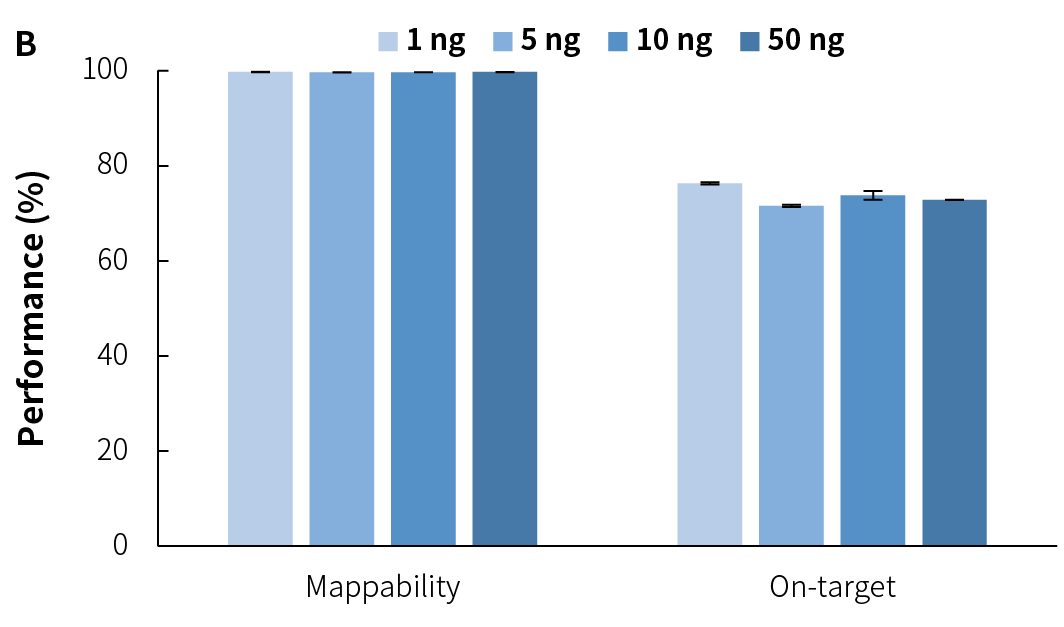
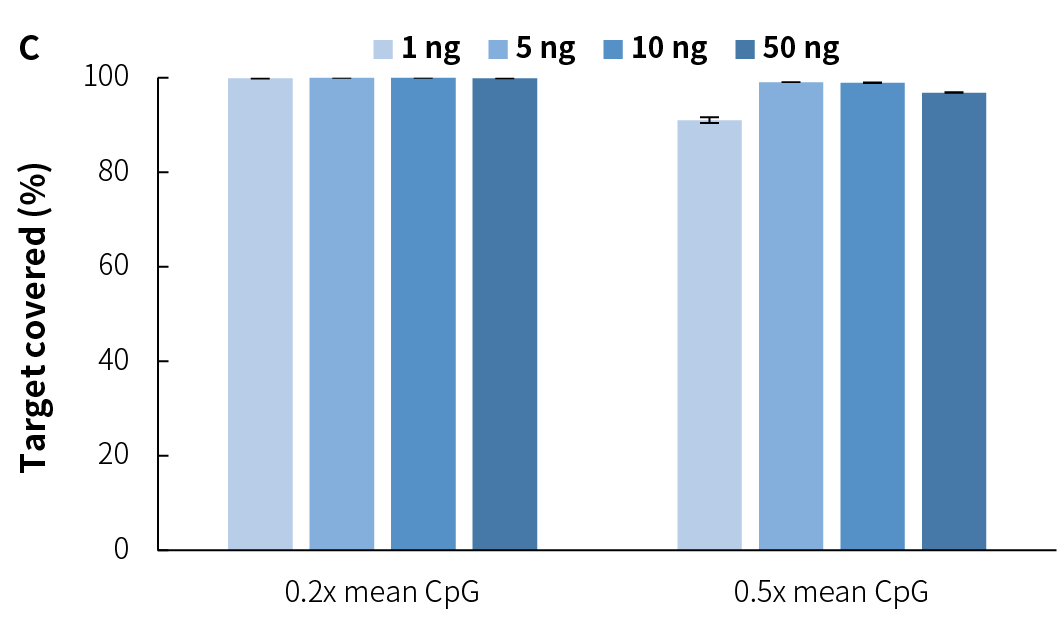
Average sequencing depth is a crucial indicator of library quality. Higher sequencing depth effectively increases data utilization, providing richer sample information, and positively influencing subsequent analysis results. Compared to v1, NadPrep v2 achieves higher effective sequencing depth across different input amounts (Figure 3.C), while maintaining high mappability and on-target (Figure 3.A), as well as target covered (Figure 3.B). Additionally, it exhibits lower GC bias (Figure 3.D), establishing a solid foundation for superior data utilization and high-quality analysis results.
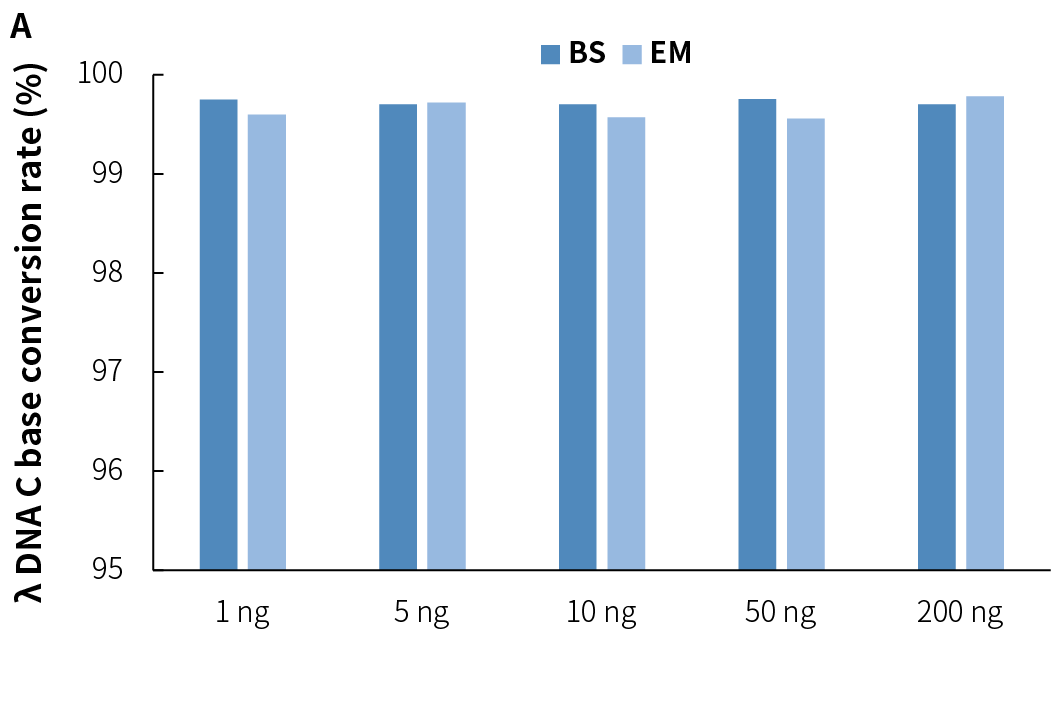
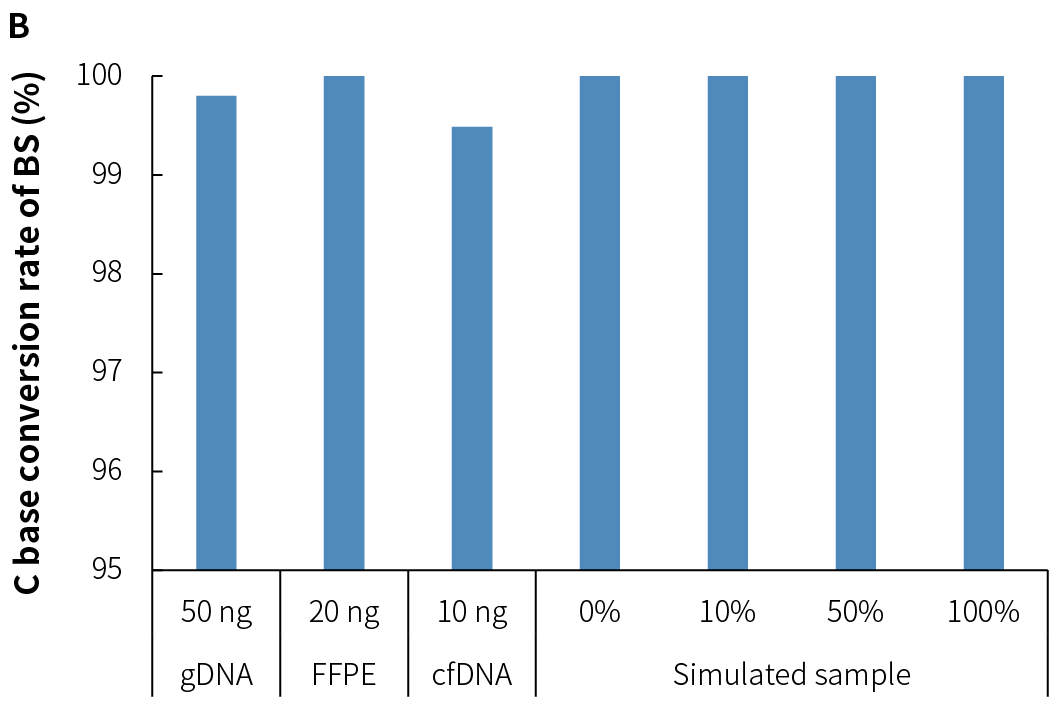
4.3 Stable Conversion Efficiency
NadPrep v2 demonstrates stable and efficient conversion rates across various sample types and input amounts when used with different conversion methods. This highlights its excellent adaptability to various application scenarios.
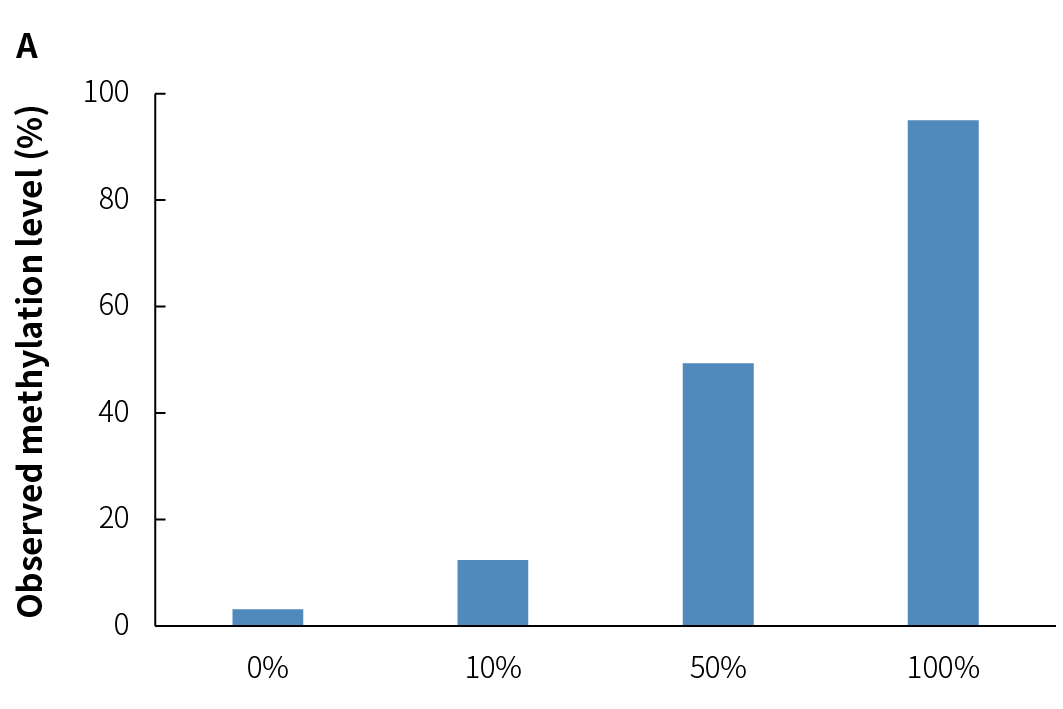
4.4 Accurate Quantification of Methylation Levels
Accurate quantification of methylation levels is a crucial measure of a sequencing solution's ability to resolve methylation frequencies at individual CpG sites, with significant implications for scientific research and clinical management. Libraries were prepared from simulated samples with varying methylation levels by using NadPrep v2 to evaluate the accuracy of methylation quantification. The results show a high correlation between detected and theoretical methylation levels (Figure 5.), demonstrating the accuracy and reliability of its methylation level quantification.
4.5 Highly Adaptability to Low Input Amounts
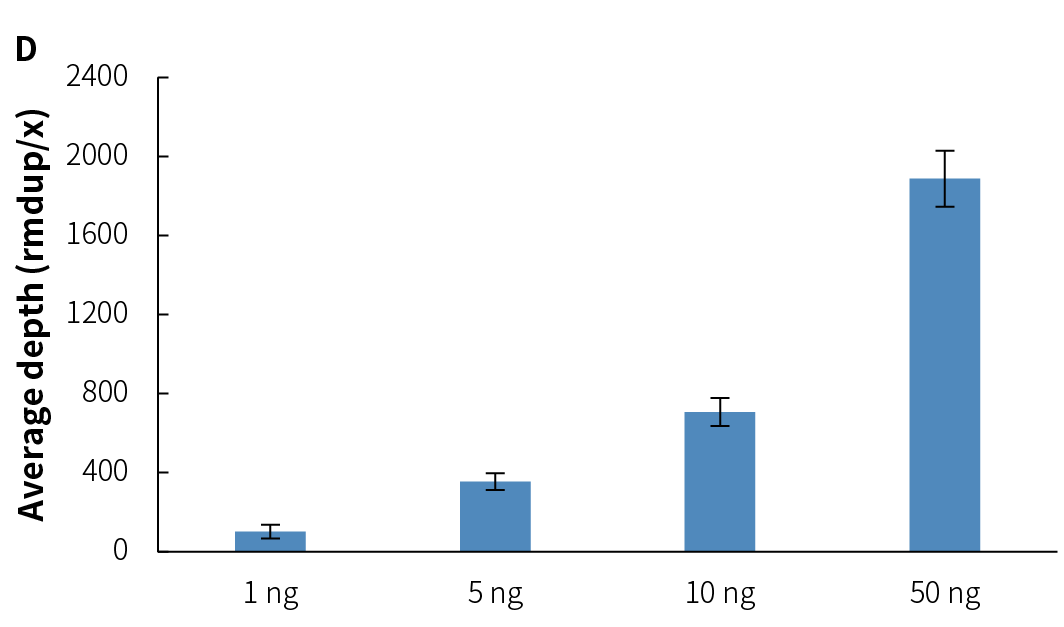
Compared to v1, NadPrep v2 lowers the input requirement from 10 ng to 1 ng, significantly enhancing its adaptability for low-input samples. After preparing the pre-library with gDNA using NadPrep v2 for ligation and EM conversion, hybrid capture was completed using Nanodigmbio's exclusive patented μCaler hybrid capture system and ultra-sensitive probes. The results demonstrate stable library yield (Figure 6.A) and capture performance (Figures 6.B & 6.C), as well as higher deduplicated average sequencing depth (Figure 6.D), effectively meeting the needs of methylation library preparation for trace or precious samples, thus providing reliable support for diverse application scenarios.
05 Conclusion
NadPrep Methyl Library Preparation Module v2 delivers outstanding library preparation performance across various sample types, ensuring efficient and stable library yield while significantly enhancing data utilization. In addition, NadPrep v2 is compatible with various methylation adapters for whole-genome methylation sequencing on Illumina and MGI platforms. Meanwhile, It can integrate seamlessly with NadPrep Hybrid Capture Reagents or the ultra-sensitive μCaler Hybrid Capture Reagents v2 for targeted methylation sequencing. With its high flexibility, NadPrep v2 is well-suited for a broad range of methylation sequencing applications, providing researchers with more options and greater efficiency.
Reference
[1] Sun Z, Behati S, Wang P, et al. Performance comparisons of methylation and structural variants from low-input whole-genome methylation sequencing[J]. Epigenomics, 2023, 15(1): 11-19.
DNA methylation, as a key epigenetic modification, holds significant value in fields such as cancer research, genetics, epigenetics, stem cell studies etc[1]. As a pivotal step in the DNA methylation sequencing workflow, the quality of methylation library preparation directly impacts the accuracy and sensitivity of detection. With the growing demands of research, the application of methylation sequencing libraries faces higher requirements in stability, precision, and efficiency, driving continuous advancements in related technologies and products to better meet the rigorous standards of scientific research and clinical applications.
02 NadPrep dsDNA Methylation Library Preparation Solution
The newly upgraded NadPrep Methyl Library Preparation Module v2 is suitable for preparing methylation sequencing (Methyl-Seq) libraries from 1-500 ng DNA samples, supporting whole-genome bisulfite sequencing (WGBS) as well as hybrid capture-based targeted methylation sequencing with effective enrichment of targeted libraries. Meanwhile, it is compatible with various types of DNA samples, including gDNA, cfDNA, and FFPE DNA, as well as a wide range of commercially available conversion solutions.
Compared to the v1 version, the upgraded module shortens experimental time, supports lower sample input amounts, and significantly improves library yield and data utilization efficiency, offering users a more efficient, flexible, and reliable methylation sequencing solution.

Figure 1. Workflow of NadPrep dsDNA Methylation Library Preparation Solution for MGI and Illumina platforms.
03 Feature
Lower Sample Input: Reduced from 10 ng to as low as 1 ng.
Faster Experimental Workflow: Shortened from 3 hr to 2.5 hr*
Higher Library Yield: Significant improvement in library yield across various applications compared to v1.
Improved Data Utilization: Achieves higher effective sequencing depth than v1, ranking among the best in its class.
Flexible Conversion Options: Supports both bisulfite conversion and enzymatic conversion methods.
Dual Platform Compatibility: Can be applied to both Illumina and MGI sequencing platforms after ligation with different adapters.
*Excluding conversion time.
04 Performance
4.1 Higher Library Yield
NadPrep Methyl Library Preparation Module v2 (hereinafter referred to as "NadPrep v2") demonstrates consistent library yield across various common sample types, including gDNA, FFPE DNA, and cfDNA (Figure 2.A), as well as simulated samples with different methylation levels (1%, 10%, 50%, and 100%) (Figure 2.B). Compared to v1, NadPrep v2 delivers significantly enhanced library yield across samples with different input amounts (Figure 2.C). This enhanced performance not only improves library yield but also supports compatibility with diverse sample types, enabling more efficient applications in diverse research scenarios.



Figure 2. NadPrep Methyl Library Preparation Module v2 ensures stable library yield for different sample types. Libraries were prepared using NadPrep Methyl Library Preparation Module v2 coupled with NadPrep Methyl Stubby Adapter (UDI) Module (with 10 nt Index) and treated by BS conversion. A. gDNA, FFPE DNA, and cfDNA; B. Simulated samples with varying methylation levels; C. gDNA (comparison of v1 vs. v2).
Note: The samples were simulated DNA by using human 100% Methylated DNA standard (Zymo, D5014-2) positive control and 0% Methylated DNA standard (Zymo, D5014-1) negative control to mimic different methylation levels (0%, 10%, and 50%), with an initial input amount of 50 ng.
4.2 Improved Data Utilization
Average sequencing depth is a crucial indicator of library quality. Higher sequencing depth effectively increases data utilization, providing richer sample information, and positively influencing subsequent analysis results. Compared to v1, NadPrep v2 achieves higher effective sequencing depth across different input amounts (Figure 3.C), while maintaining high mappability and on-target (Figure 3.A), as well as target covered (Figure 3.B). Additionally, it exhibits lower GC bias (Figure 3.D), establishing a solid foundation for superior data utilization and high-quality analysis results.
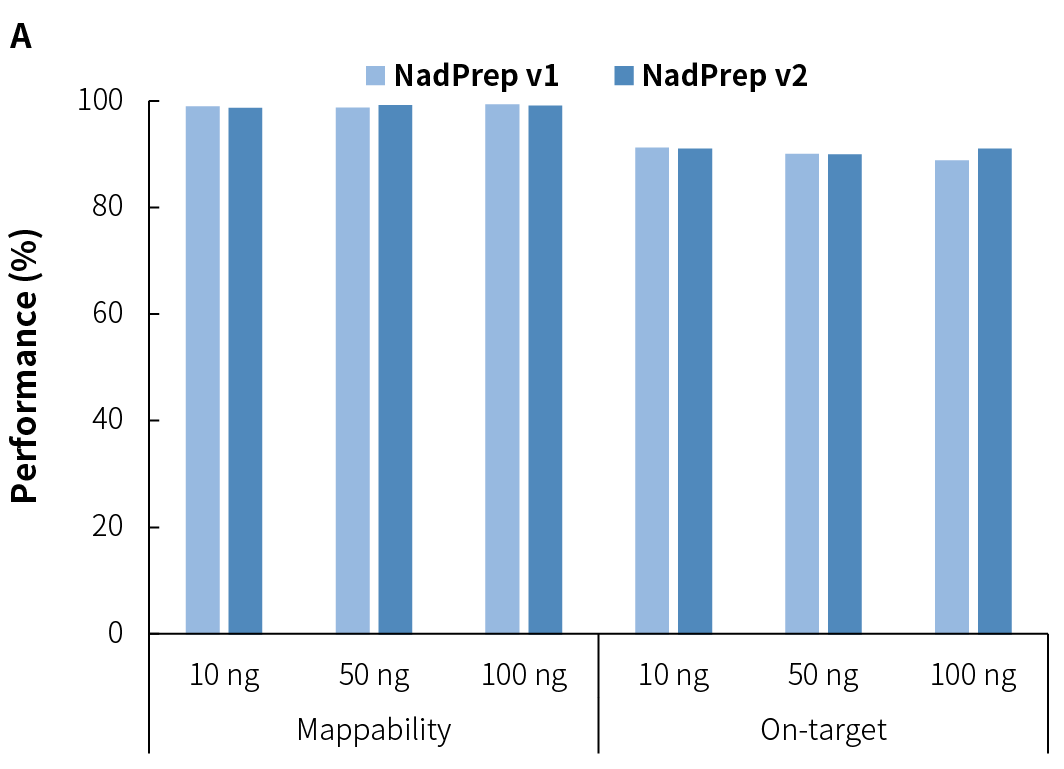
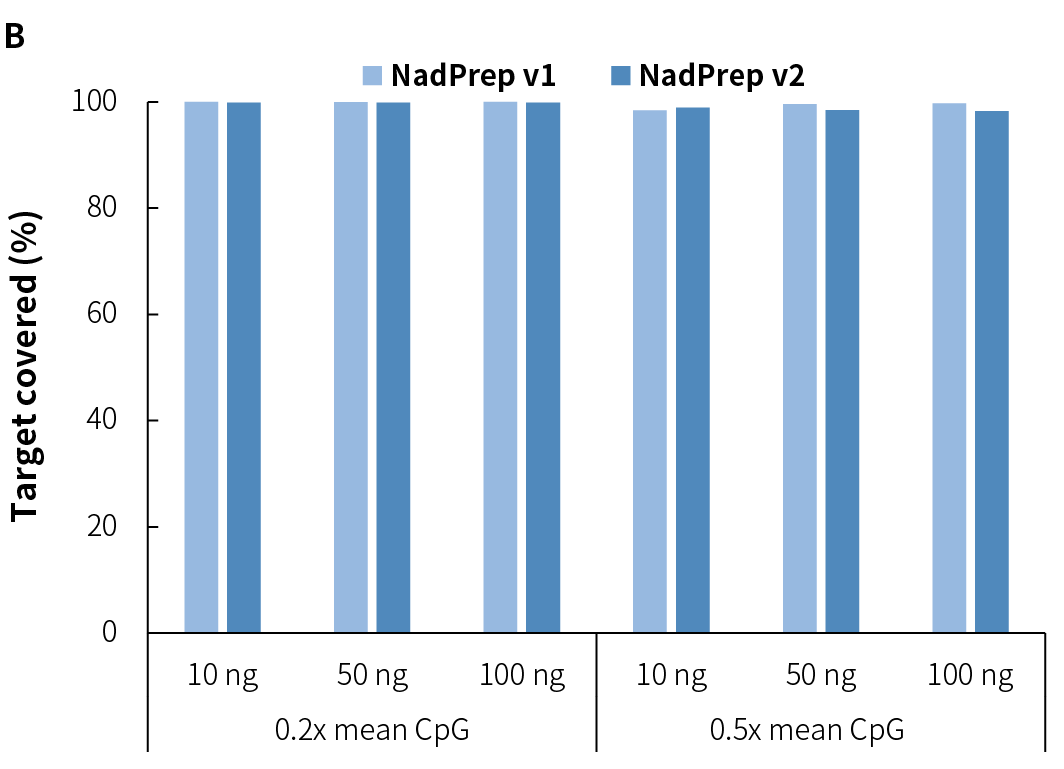

4.3 Stable Conversion Efficiency
NadPrep v2 demonstrates stable and efficient conversion rates across various sample types and input amounts when used with different conversion methods. This highlights its excellent adaptability to various application scenarios.
4.4 Accurate Quantification of Methylation Levels
Accurate quantification of methylation levels is a crucial measure of a sequencing solution's ability to resolve methylation frequencies at individual CpG sites, with significant implications for scientific research and clinical management. Libraries were prepared from simulated samples with varying methylation levels by using NadPrep v2 to evaluate the accuracy of methylation quantification. The results show a high correlation between detected and theoretical methylation levels (Figure 5.), demonstrating the accuracy and reliability of its methylation level quantification.
4.5 Highly Adaptability to Low Input Amounts
Compared to v1, NadPrep v2 lowers the input requirement from 10 ng to 1 ng, significantly enhancing its adaptability for low-input samples. After preparing the pre-library with gDNA using NadPrep v2 for ligation and EM conversion, hybrid capture was completed using Nanodigmbio's exclusive patented μCaler hybrid capture system and ultra-sensitive probes. The results demonstrate stable library yield (Figure 6.A) and capture performance (Figures 6.B & 6.C), as well as higher deduplicated average sequencing depth (Figure 6.D), effectively meeting the needs of methylation library preparation for trace or precious samples, thus providing reliable support for diverse application scenarios.

05 Conclusion
NadPrep Methyl Library Preparation Module v2 delivers outstanding library preparation performance across various sample types, ensuring efficient and stable library yield while significantly enhancing data utilization. In addition, NadPrep v2 is compatible with various methylation adapters for whole-genome methylation sequencing on Illumina and MGI platforms. Meanwhile, It can integrate seamlessly with NadPrep Hybrid Capture Reagents or the ultra-sensitive μCaler Hybrid Capture Reagents v2 for targeted methylation sequencing. With its high flexibility, NadPrep v2 is well-suited for a broad range of methylation sequencing applications, providing researchers with more options and greater efficiency.
Reference
[1] Sun Z, Behati S, Wang P, et al. Performance comparisons of methylation and structural variants from low-input whole-genome methylation sequencing[J]. Epigenomics, 2023, 15(1): 11-19.
Solutions
- Methyl Library Preparation Total Solution
- Sequencing single library on different platform--Universal Stubby Adapter (UDI)
- HRD score Analysis
- Unique Dual Index for MGI platforms
- RNA-Cap Sequencing of Human Respiratory Viruses Including SARS-CoV-2
- Total Solution for RNA-Cap Sequencing
- Total Solution for MGI Platforms
- Whole Exome Sequencing
- Low-frequency Mutation Analysis
Events
-
Exhibition Preview | Nanodigmbio invites you to join us at Boston 2025 Annual Meeting of the American Society of Human Genetics (ASHG)

-
Exhibition Preview | Nanodigmbio Invites You to Join Us at WHX & WHX Labs Kuala Lumpur 2025, Malaysia International Trade and Exhibition Centre in Kuala Lumpur

-
Exhibition Preview | Nanodigmbio Invites You to Join Us at Hospitalar 2025, Brazil International Medical Device Exhibition in São Paulo

-
Exhibition Preview | Nanodigmbio invites you to join us at Denver 2024 Annual Meeting of the American Society of Human Genetics (ASHG)

-
Exhibition Preview | Nanodigmbio invites you to join us at Sapporo 2024 Annual Meeting of the Japan Society of Human Genetics (JSHG)

-
Exhibition Preview | Nanodigmbio invites you to join us at Association for Diagnostics & Laboratory Medicine (ADLM)